GENtle/Automatic annotation
< GENtle
The automatic annotation feature can search a database of standard vectors (included with the GENtle package), and (optionally) a user-generated database, for feature sequences that are found in the currently opened DNA sequence. Recognized features are then annotated in the current sequence.
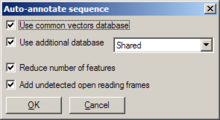
Invoked through Edit/Auto-annotate sequence or F9, a dialog opens, offering various settings:
- Whether or not to search the common vectors database
- Whether or not to use a user-generated database (and, if so, which one)
- Whether or not to reduce the number of generated features (recommended; otherwise, a lot of features are annotated)
- Whether or not to add unrecognized open reading frames as features