Biochemistry/DNA and RNA
Deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) are the information storage molecules and working templates for the construction of proteins. Every living cell and virus encodes its genetic information using either DNA or RNA. It is a true marvel of evolution that the vast amount of information needed to produce a human being can fit inside cells.
Friedrich Miescher first isolated DNA and RNA from used surgical bandages in 1869. A series of experiments done by Oswald Avery, Colin MacLeod, and Maclyn McCarty in 1944 defined DNA as the genetic information. They injected virulent encapsulated bacteria into a mouse. As it a result, it died. On the contrary, nonvirulent nonencapsulated bacteria did not kill the mouse. Also, heating the virulent encapsulated bacteria and injecting did not kill the mouse either. The surprise lied in mixing the heat-killed virulent encapsulated bacteria with nonvirulent nonencapsulated bacteria did kill the mouse. Avery's group kept isolating factors until they discovered what killed the mouse, which was the DNA. In conclusion, the nonpathogenic bacteria was made pathogenic by DNA transfer from a pathogenic stain. A.D. Hershey and Martha Chase's work in 1952 reconfirmed that DNA is the carrier of genetic information. Hershey and Chase's group conducted an experiment involving two phages. One of the phages was radioactively labeled with heavy phosphorus to highlight the DNA and heavy sulfur to highlight the protein. The two phages inserted their "genetic material" into the bacterial cell. Afterwards, these cells were "blended" to remove the phage heads. The resulting radioactively-labeled heavy phosphorus was found in the cell. This experiment showed that DNA alone was sufficient to make new viruses, and therefore, defined as the genetic information. Rosalind Franklin, Chargaff, and other chemists provided information that led to the discovery of DNA as well. Franklin provided crystallographic data that implied a helical structure. Chargaff's rule said that the adenine content was equal to the thymine content and cytosine was equal to guanine. Also the aromaticity of the electron-rich purine and pyrimidine rings allowed for tautomer shifts between the keto and enol form. The keto tautomer (lactam) dominated at physiological pH. Garnering all this information, James Watson and Francis Crick discovered the structure of DNA (double-helix) in 1953.
Structure of DNA and RNA
editDNA is a polymer of nucleotides. Nucleotides consist of a pentose sugar (2-deoxyribose for DNA and ribose for RNA), a phosphate group (), and a nitrogen-containing base. There are five nitrogen bases - adenine, guanine, thymine, cytosine and uracil. Individual nucleotides are connected by phosphodiester bonds to form polynucleotides.
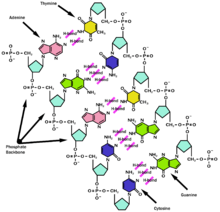
DNA exists as a double helix of two (2) strands of polynucleotides. According to the principle of complementarity nucleotides A (adenine) bases are bound with a hydrogen bridge to the T (thymine) or U (uracil in case of RNA) on the other thread and similarly C (cytosine) bind themselves to G (guanine). This principle allows for DNA and RNA replication and for limited possibilities of repairing the genetic information when one of the threads gets damaged.
Difference between DNA and RNA
editDNA is the permanent genetic information storage medium, found in the nucleus of most cells of most living organisms. RNA is, in the case of eukaryotes, the medium that transfers the genetic information from the nucleus to the cytoplasm where proteins are synthesized. The most basic structural difference between a DNA molecule and an RNA molecule is that DNA lacks an hydroxyl group at its 2' carbon while RNA has an hydroxyl group at its 2' carbon.
There are three major types of RNA:
- messenger RNA (mRNA) - temporarily created RNA used for transferring the information from the DNA in the nucleus to the ribosome in the cytoplasm where it is "read," or translated, and a protein is synthesized.Each gene produce a separate mRNA molecule when a certain protein is needed in the cell. In addition, the size of an mRNA depends on the number of nucletides in that particular gene.
- transfer RNA (tRNA) - Is the smallest of the RNA molecules and it is associated with the ribosome and delivers an amino acid to be attached to the growing protein. Only the tRna can translate the genetic information into amino acid for proteins.
Each tRNA contains an "anticodon" which is a series of 3 bases that complements 3 bases on mRNA.
- ribosomal RNA (rRNA) - a major constituent of the ribosome with both structural and catalytic properties.
Additionally, by 2005, other types of RNA, such as siRNA (small, interfering RNA) and miRNA have been characterized.
DNA structure is typically a double stranded molecule with long chain of nucleotides, while RNA is a single stranded molecule " in most biological roles" and it has shorter chain of nucleotide. Deoxyribose sugar in DNA is less reactive because of the carbon- hydrogen bonds. DNA has small grooves to prevent the enzymes to attack DNA. on the other hand, RNA contain ribose sugar which is more reactive because of the carbon-hydroxyl bonds. RNA not stable in its alkaline conditions.RNA has larger grooves than that of DNA, which makes RNA easier to be attacked by enzymes. DNA has a unique features because of its helix geometry which is in B-form. DNA is protected completely by the body in which the body destroys any enzymes that cleave DNA. In contrast the helix geometry of RNA is in the A- form; RNA can be broken down and reused. RNA is more resistance to be damage by Ultra- violet rays, while DNA can be damaged if exposed to Ultra-violet rays. DNA can be found in nucleus, while RNA can be found in nucleus and cytoplasm. The bases of DNA are A,T,C,and G, DNA is a long polymer with phosphate and deoxyribose backbone. In contrast, RNA bases are A,U, C,and G, RNA backbone contain ribose and phosphate. The job of DNA on the cell is to store and transmit genetic information. RNA job is to transfer the genetic codes that are needed for the creation of proteins from the nucleus to the ribosome, this process prevents the DNA from leaving the nucleus, so DNA stays safe.
Plasmid
editPlasmids are small, circular DNA molecules that replicate separately from the much larger bacterial chromosome can be use in laboratory to manipulate genes. Plasmids can carry almost any gene and they can passed it on from one generation of bacteria to the next generation, therefore plasmids are key tools for “gene cloning” which is the production of multiple identical copies of a gene carrying piece of DNA
DNA Replication ;Semi-conservative replication
editIn the process of DNA replication, as cells divide, copies of DNA must be produced in order to transfer the genetic information. In DNA replication, the strands in the original DNA separate, and then each of the parent strands make copies by synthesizing complementary strands. Enzyme called helicase will start The process of replication by catalyzing the unwinding of protein of the double helix. This enzyme can break the H bonds between the complementary bases. These single strands now act as templates for the synthesis of new complementary strands. The energy for the reaction can be provided when a nucleoside triphosphate bonds to a sugar (at the end of a growing strand), and 2 phosphate group are cleaved. Then a T in the template forms H bond with an A in ATP, and a G on the template forms H bond with CTP. After the entire double helix of the parent DNA is copied, the new DNA molecule will form . The new DNA molecule is consist of one strand from the parent (original) DNA and one strand from the daughter strand( newly synthesized strand). This process is called "semi-conservative replication".
Recombination
editRecombination refers to ways in which DNA modification leads to distinct gene expressions and functions. Deletions, insertions and substitutions are the most useful changes implemented for the synthesis of new genes. Recombination techniques and recombinant DNA technologies has made possible the modification and creation of new genes for specific proposes. These techniques helps clone, amplified and introduce new DNA into suitable cells for replication. Restriction enzymes and DNA ligase play a vital role in the production of recombinant DNA. Vectors are useful for cloning. Plasmids or viral DNA (lambda Phage) used to introduced a DNA sequence into a cell is refer as vector.
The semi-conservative hypothesis was confirmed by both Matthew Meselson and Franklin Stahl in 1958. The parent DNA was labeled with "heavy nitrogen," the radioisotope of fifteen. This radioactive nitrogen was made through growing E. coli with ammonium chloride (also with "heavy nitrogen") environment. Directly after the labeling of "heavy nitrogen" to the parent DNA strand, it was transferred to a medium with only regular nitrogen (isotope of fourteen). The proposed question was "What is the distribution between the two nitrogens in the DNA molecules after successive rounds of replication?" Using density-gradient equilibrium sedimentation, this question was answered. In this centrifugation, similar densities were grouped together in the tube. Afterwards, an absorbance was applied with ultra-violet light to determine the density bands of all the different kinds of DNA in the solution. After one generation, a unique single density band was shown. It was not exactly the band of heavy nitrogen nor the regular band of regular nitrogen, but rather halfway between the two. This proved that DNA was not conservative but semi-conservative due to this nitrogen hybrid. One generation after this one, there were two nitrogen-14 molecules and two hybrid nitrogen molecules. The probability and statistics of this second generation reassures the semi-conservative hypothesis.
As DNA strands can combine and form molecules, it can also be melted, resulting in separation or denaturation. It is important to note that the heat from the melting process does not disrupt the DNA structure, but rather DNA helicase takes energy from the heat, separating the DNA strands. The denaturation process occurs at about ninety-four degrees Fahrenheit where the double strands are converted to single stands. Absorbance readings were taken to reconfirm the denaturation process. When the DNA molecule is in its double-stranded form, it absorbs less UV light. In its single-stranded form, it absorbs more UV light because the single strand is more exposed than its double-strand counterpart. This is known as the hypochromic effect. In a PCR, polymerase chain reaction, it utilizes this denaturation capability in its primary step. Afterwards, it is then annealed at fifty to sixty degrees Fahrenheit. In this step, this is where the primers bind to their corresponding flanking DNA base sequence. In the last step, DNA polymerization, it requires a few ingredients: heat-resistant DNA polymerase, magnesium ion, buffer, and corresponding nucleotides. The DNA polymerase requires heat resistance because throughout the entire process, it undergoes a constant temperatures changes. The magnesium stabilizes the phosphate backbone. The buffer prevents drastic changes in pH, because if the pH lies near the extremes on the scale, it may denature the DNA so drastically that it will not recombine. This final step of DNA polymerization happens at the "melting temperature." The melting temperature is the temperature at which half of the DNA is single-stranded and the other half is double-stranded. At this optimal temperature of seventy-two degrees Fahrenheit, the DNA is amplified with its respective primers, replicating at an exponential rate.
DNA manipulation: human's effort to desire DNA and "read" DNA
editRestriction endonucleases
editWhat's endonuclease?--Endonucleases are enzymes that can recognize specific base sequences in double-helical DNA and cleave, at specific places, both strands of that duplex. Many prokaryotes contain restriction enzymes. Biologically, they cleave foreign DNA molecules. Because the sites recognized by its own restriction enzymes are metylated, the cells' own DNA are not cleaved. One striking properties of endonucleases is that they recognize DNA cleavage sites by "two-fold symmetry". In another word, within the cleavage site domain (usually 4 to 8 base pair), there exists a center, around which the cleavage site rotate 180 degree, the whole cleavage site would be indistinguishable. The following graph shows an example of how endonuclease recognize cleavage site by symmetry..After cleavage from several different endonulcease, a DNA sample would be sliced into many different duplex fragments, which are then separated by electrophoresis. The restriction fragments are then denatured to form single stranded DNA, which are then transferred to a nitrocellulose sheet, where they are exposed to a 32P-labeled single-stranded DNA probe. The single-stranded DNA are then hybridized by the radio active complementary strand. These duplex are then examined by autoradiography, which reveals the sequence of the DNA. This technique is named Southern plot, after Edwin South, who invented the technique. The following image shows the steps for creating a southern plot.
DNA sequencing
editDNA subunits are dGMP, dAMP, dCMP and dTMP. In a DNA molecule, these subunits are connected in a way that their phosphate on 5 prime carbon are bonded to another subunit's hydroxy group on 3 prime carbon. If there is no hydroxy group on 3 prime carbon, then the DNA polymeration will be terminated. Utilizing this idea, subunit analogies were invented, ddGMP, ddAMP, ddCMP and ddTMP. These analogies have the same structure as their parent molecule except that they have hydrogen on 3 prime carbon instead of hydroxyl group. In practice, target DNA template is put into four polymeration environments, dNTP+ddGMP or ddAMP or ddCMP or ddTMP, respectively.. Statically, fragments with different length terminated at base, G, A, C, T will be formed, respectively. Then these fragments will be separated by electrophoresis, which will tell the sequence of the target DNA.
DNA synthesis
editAlthough nature synthesize DNA from 5 prime carbon to 3 prime carbon, human synthesize DNA from 3 prime carbon to 5 prime carbon. The following monomer is commercially available for each of the base and is used as basic building block for DNA synthesis. It starts with the subunit with the first base bonded to resin by the 3 prime hydroxyl group. The second monomer with its own 5 hydroxyl protected by DMT (protecting group) was activated on its 3 prime phosphate, which is attached to the 5 prime hydroxyl group by nucleophilic substitution. Then the phosphite is oxidized by iodine to yield phosphate. Then DMT is removed to make the 5 prime hydroxyl available for the incoming of the third monomer. The graphical representation is shown below.
Polymerase Chain Reaction (PCR)
editIt is used for amplification of DNA. It includes three stages: DNA denaturation, oligonucleotide annealing, and DNA polymerization. In each cycle, the DNA duplex is denatured at first, then the primers for both strands (forward and reverse) bind to the desired sites and at last both strands polymerize to form the desired sequence. What needed in PCR are the DNA containing the desired sequence, primers for both strands, heat resistant DNA polymerase, dNTP and constantly changing temperature. The following image shows the detailed steps in PCR.
RNA Editing
editIntroduction
editAdenosine deaminases (ADAR) are mRNA editing enzymes that alternate a double stranded RNA sequence by tuning it. When tuning the mRNA, ADARs retype the nucleotides of the mRNA at certain, specific points they wish to change. Because ADARs can change the sequence of mRNA, they can create or remove a splice site, delete or alter the meaning of a codon, and finally change the sequence of the RNA altogether. ADAR plays a vital part in editing mRNA and modulating the mRNA translational activity and is mainly found in the nervous system and is important in regulating the nervous systems. Lacking the ADAR gene is very detrimental to health.
Function
edit
RNA splicing and RNA editing are very similar to one another. RNA splicing is more of a cut, copy, and past process while RNA editing is an alteration of one or more nucleotides. There are mainly two types of ADAR that alternate the sequence of a single nucleotide. The first type of ADAR retypes cytidine nucleotides into uridine nucleotides while a second type of ADAR retypes adenosine into inosine.
mRNA consisting of many inosine nucleotides in its sequence is known to be very abundant in the nucleus and only mRNAs that leave the nucleus can be translated. In this case, ADAR is modulating the translational activities of mRNAs. In modulating the translational activities of the specified mRNA, it is also modulating the abundance of the certain proteins that are expressed in the edited mRNA. Also, increasing inosine can stabilize the structure of mRNA. Hence, ADAR can function as an mRNA stabilizer too.
ADARs target the coding sequence, introns, and 5’ and 3’ untranslated reigions (UTR). ADAR targets the coding sequence by changing the nucleotide which then changes the geometry of the protein. Change in the geometry of the protein may yield to higher protein-protein affinity interaction. This high protein-protein affinity can improve biological reactions by facilitating those reactions. Alternation of the non-coding sites such as introns, 5’, and 3’ UTR is still unknown. The only thing that is known is that alternating the sequence within these sites can create new splice sites.
Importance of ADAR
edit
Adar is mainly found in the nervous system because it plays a very important role in regulating the nervous system. One of Adars’ primary functions is its regulation of the nervous system. Adar regulates the nervous system by editing the glutamate receptors’ mRNA. These glutamate receptors construct the glutamate-gated ion channels which are important in transferring electrical signals between neurons. When ADARs edit gluR mRNA, they change the calcium permeability of these glutamate channels. They can either lower the calcium permeability for certain glutamate channel or increase calcium permeability for other glutamate channels. Regulation of calcium permeability is important to proper neurotransmission between neurons.
Disease connecting to ADAR
edit
Adenosine deaminase Deficiency (ADD) is caused by the lack of ADAR gene. Hence ADARs editing is important for survival especially for human. Without ADAR, the body can not break down the toxin deoxyadenosine. The accumulation of this toxin destroys the immune cells making its host vulnerable to infection from bacteria and viruses.
Seizures can also be caused by ADAR. Unedited version of gluR mRNA strongly causes seizures. When a seizure happens, the body mechanism increases the level of ADAR activity to edit gluR mRNA to prevent future seizures.
DNA Replication:Initiation
edit
There are multiple mechanisms that have been proposed for the melting of the DNA strand and the subsequent unwinding of the double helix during the initiation process of DNA replication.
Two recently proposed models are E1 and LTag. These two models function differently to attain the same overall goal of melting the DNA double helix due to differences in their overall structure.
E1 model: In the E1 hexamer, there are six ß-hairpins in the central channel of the protein. They are arranged in a staircase-like structure. Two adjacent E1 trimers assemble at the origin point to melt the double-stranded DNA. A ring shaped E1 hexamer is then formed around the melted single-stranded DNA and the DNA is then pumped through the ring hexamer to from a fork that allows for the replication of the DNA.
Ltag: The channel diameters of these hexamers vary between 13-17 angstroms. The hexamer responsible for melting has an assortment of planar ß-hairpins in the narrow channel. Two of these hexamers surround the origin point of the double-stranded DNA and squeeze the area together. This causes the melting of the dsDNA by forcing the breakage of base pairing. The two single-stranded DNA strands are then pumped into a larger channeland finally out through two separate Zn-domains. This allows for the replication of the DNA.
The two mechanisms here are built solely on the structural knowledge of the proteins involved in the process and as such more experimental support is needed to confirm.
There are not an excessive amount of helicases. Currently, work is being done in discerning the more simple helicases, and trying to find the different mechanisms that they may have. While there are some similarities, for example helicases all seem to have a hexameric structure, there are also differences as well in their structures that contribute greatly to the differences in their mechanisms.
-This article refers to a recently published article in Curr Opin Struct Biol. published 2010, Sep. 24.: "Origin DNA melting and unwinding in DNA replication" by Gai et al.
