An Introduction to Molecular Biology/Gene Expression
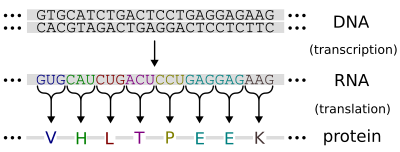
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as ribosomal RNA (rRNA) genes or transfer RNA (tRNA) genes, the product is a functional RNA. The process of gene expression is used by all known life - eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea) and viruses - to generate the macromolecular machinery for life. Several steps in the gene expression process may be modulated, including the transcription, RNA splicing, translation, and post-translational modification of a protein. Gene regulation gives the cell control over structure and function, and is the basis for cellular differentiation, morphogenesis and the versatility and adaptability of any organism. Gene regulation may also serve as a substrate for evolutionary change, since control of the timing, location, and amount of gene expression can have a profound effect on the functions (actions) of the gene in a cell or in a multicellular organism.
In genetics, gene expression is the most fundamental level at which genotype gives rise to the phenotype. The genetic code stored in DNA in form of nucleotide sequence is "interpreted" by gene expression, and the properties of the expression products give rise to the organism's phenotype.[1] A molecule which allows the genetic material to be realized as a protein was first hypothesized by François Jacob and Jacques Monod. RNA synthesis by RNA polymerase was established in vitro by several laboratories by 1965; however, the RNA synthesized by these enzymes had properties that suggested the existence of an additional factor needed to terminate transcription correctly. In 1972, Walter Fiers became the first person to actually prove the existence of the terminating enzyme. Roger D. Kornberg won the 2006 Nobel Prize in Chemistry "for his studies of the molecular basis of eukaryotic transcription.
Transcription
editGeneration of RNA from DNA is known as transcription. In other word transcription is the process of creating a complementary RNA copy of a sequence of DNA. During transcription, a DNA sequence is read by RNA polymerase, which produces a complementary, antiparallel RNA strand. As opposed to DNA replication, transcription results in an RNA complement that includes uracil (U) in all instances where thymine (T) would have occurred in a DNA complement.[2] Transcription can be explained easily in 4 or 5 simple steps, each moving like a wave along the DNA.
As the Hydrogen Bonds Break DNA unwinds. The free nucleotides of the RNA, pair with complementary DNA bases. RNA sugar-phosphate backbone forms. (by RNA Polymerase.) Hydrogen bonds of the untwisted RNA-DNA "ladder" break, freeing the new RNA. The RNA is further processed and then moves through the small nuclear pores to the cytoplasm.
Transcription is the first step leading to gene expression. The stretch of DNA transcribed into an RNA molecule is called a transcription unit and encodes at least one gene. If the gene transcribed encodes a protein, the result of transcription is messenger RNA (mRNA), which will then be used to create that protein via the process of translation. Alternatively, the transcribed gene may encode for either ribosomal RNA (rRNA) or transfer RNA (tRNA), other components of the protein-assembly process, or other ribozymes.
A DNA transcription unit encoding for a protein contains not only the sequence that will eventually be directly translated into the protein (the coding sequence) but also regulatory sequences that direct and regulate the synthesis of that protein. The regulatory sequence before (upstream from) the coding sequence is called the 5'UTR (five prime untranslated region) , and the sequence following (downstream from) the coding sequence is called the 3'UTR (three prime untranslated region) . Transcription has some proofreading mechanisms, but they are fewer and less effective than the controls for copying DNA; therefore, transcription has a lower copying fidelity than DNA replication. As in DNA replication, DNA is read from 3' → 5' during transcription. Meanwhile, the complementary RNA is created from the 5' → 3' direction. This means its 5' end is created first in base pairing. Although DNA is arranged as two antiparallel strands in a double helix, only one of the two DNA strands, called the template strand, is used for transcription. This is because RNA is only single-stranded, as opposed to double-stranded DNA. The other DNA strand is called the coding strand, because its sequence is the same as the newly created RNA transcript (except for the substitution of uracil for thymine). The use of only the 3' → 5' strand eliminates the need for the Okazaki fragments seen in DNA replication. Transcription is divided into 5 stages: pre-initiation, initiation, promoter clearance, elongation and termination.
One gene-one enzyme hypothesis
editThe one gene-one enzyme hypothesis is the idea that genes act through the production of enzymes, with each gene responsible for producing a single enzyme that in turn affects a single step in a metabolic pathway. The concept was proposed by George Beadle and Edward Tatum in an influential 1941 paper on genetic mutations in the mold Neurospora crassa,[3] and subsequently was dubbed the "one gene-one enzyme hypothesis" by their collaborator Norman Horowitz. It is often considered the first significant result in what came to be called molecular biology. Although it has been extremely influential, the hypothesis was recognized soon after its proposal to be an oversimplification. Even the subsequent reformulation of the "one gene-one polypeptide" hypothesis is now considered too simple to describe the relationship between genes and proteins.[4]
What is Neurospora? Neurospora crassa is a type of red bread mold of the phylum Ascomycota. The genus name, meaning "nerve spore" refers to the characteristic striations on the spores.
N. crassa is used as a model organism because it is easy to grow and has a haploid life cycle that makes genetic analysis simple since recessive traits will show up in the offspring. Analysis of genetic recombination is facilitated by the ordered arrangement of the products of meiosis in Neurospora ascospores. Its entire genome of seven chromosomes has been sequenced. Neurospora was used by Edward Tatum and George Wells Beadle in their experiments for which they won the Nobel Prize in Physiology or Medicine in 1958. Beadle and Tatum exposed N. crassa to x-rays, causing mutations. They then observed failures in metabolic pathways caused by errors in specific enzymes. This led them to propose the "one gene, one enzyme" hypothesis that specific genes code for specific proteins. Their hypothesis was later elaborated to enzyme pathways by Norman Horowitz, also working on Neurospora.
One gene-one polypeptide
editBy the early 1950s, advances in biochemical genetics—spurred in part by the original hypothesis—made the one gene-one enzyme hypothesis seem very unlikely (at least in its original form). Beginning in 1957, Vernon Ingram and others showed through protein fingerprinting that genetic variations in proteins (such as sickle cell hemoglobin) could be limited to differences in just a single polypeptide chain in a multimeric protein, leading to a "one gene-one polypeptide" hypothesis instead. According to geneticist Rowland H. Davis, "By 1958 – indeed, even by 1948 – one gene, one enzyme was no longer a hypothesis to be resolutely defended; it was simply the name of a research program." Presently, the one gene-one polypeptide perspective cannot account for the various spliced versions in many eukaryote organisms which use a spliceosome to individually prepare a RNA transcript depending on the various inter- and intra-cellular environmental signals. This splicing was discovered in 1977 by Phillip Sharp and Richard J. Roberts.
Operon
editAn operon is a functioning unit of genomic material containing a cluster of genes under the control of a single regulatory signal or promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo trans-splicing to create monocistronic mRNAs that are translated separately, i.e. several strands of mRNA that each encode a single gene product. The result of this is that the genes contained in the operon are either expressed together or not at all. Several genes must be both co-transcribed and co-regulated to define an operon. Originally operons were thought to exist solely in prokaryotes but since the discovery of the first operons in eukaryotes in the early 1990s, more evidence has arisen to suggest they are more common than previously assumed.
Operons occur primarily in prokaryotes but also in some eukaryotes, including nematodes such as C. elegans. , and Drosophila melanogaster flies. rRNA genes often exist in operons that have been found in a range of eukaryotes including chordates. An operon is made up of several structural genes arranged under a common promoter and regulated by a common operator. It is defined as a set of adjacent structural genes, plus the adjacent regulatory signals that affect transcription of the structural genes. The regulators of a given operon, including repressors, corepressors, and activators, are not necessarily coded for by that operon. The location and condition of the regulators, promoter, operator and structural DNA sequences can determine the effects of common mutations. Operons are related to regulons, stimulons and modulons. Whereas operons contain a set of genes regulated by the same operator, regulons contain a set of genes under regulation by a single regulatory protein, and stimulons contain a set of genes under regulation by a single cell stimulus.[5]
Structure of an operon
editPromoter – a nucleotide sequence that enables a gene to be transcribed. The promoter is recognized by RNA polymerase, which then initiates transcription. In RNA synthesis, promoters indicate which genes should be used for messenger RNA creation – and, by extension, control which proteins the cell manufactures.
Operator – a segment of DNA that a regulator binds to. It is classically defined in the lac operon as a segment between the promoter and the genes of the operon. In the case of a repressor, the repressor protein physically obstructs the RNA polymerase from transcribing the genes.
Structural genes – the genes that are co-regulated by the operon.
Prokaryotic promoters
editIn prokaryotes, the promoter consists of two short sequences at -10 and -35 positions upstream from the transcription start site. Sigma factors not only help in enhancing RNAP binding to the promoter but also help RNAP target specific genes to transcribe. The sequence at -10 is called the Pribnow box, or the -10 element, and usually consists of the six nucleotides TATAAT. The Pribnow box is absolutely essential to start transcription in prokaryotes. The other sequence at -35 (the -35 element) usually consists of the seven nucleotides TTGACAT. Its presence allows a very high transcription rate. Both of the above consensus sequences, while conserved on average, are not found intact in most promoters. On average only 3 of the 6 base pairs in each consensus sequence is found in any given promoter. No promoter has been identified to date that has intact consensus sequences at both the -10 and -35; artificial promoters with complete conservation of the -10/-35 hexamers has been found to promote RNA chain initiation at very high efficiencies. Some promoters contain a UP element (consensus sequence 5'-AAAWWTWTTTTNNNAAANNN-3'; W = A or T; N = any base) centered at -50; the presence of the -35 element appears to be unimportant for transcription from the UP element-containing promoters. It should be noted that the above promoter sequences are only recognized by the sigma-70 protein that interacts with the prokaryotic RNA polymerase. Complexes of prokaryotic RNA polymerase with other sigma factors recognize totally different core promoter sequences.
<-- upstream downstream -->
5'-XXXXXXXPPPPPXXXXXXPPPPPPXXXXGGGCCGGGTTGGTTGGGCCGAAGGGTTGGCCGGGGGGGGXXXX-3'
-35 -10 Gene to be transcribed
Eukaryotic promoters
editEukaryotic promoters are extremely diverse and are difficult to characterize. They typically lie upstream of the gene and can have regulatory elements several kilobases away from the transcriptional start site(enhancers). In eukaryotes, the transcriptional complex can cause the DNA to bend back on itself, which allows for placement of regulatory sequences far from the actual site of transcription. Many eukaryotic promoters, between 10 and 20% of all genes contain a TATA box (sequence TATAAA), which in turn binds a TATA binding protein which assists in the formation of the RNA polymerase transcriptional complex. The TATA box typically lies very close to the transcriptional start site (often within 50 bases).
Eukaryotic promoter regulatory sequences typically bind proteins called transcription factors which are involved in the formation of the transcriptional complex. An example is the E-box (sequence CACGTG), which binds transcription factors in the basic-helix-loop-helix (bHLH) family (e.g. BMAL1-Clock, cMyc).
Enhancer
editAn enhancer is a short region of DNA that can be bound with proteins (namely, the trans-acting factors, much like a set of transcription factors) to enhance transcription levels of genes (hence the name) in a gene cluster. While enhancers are usually cis-acting, an enhancer does not need to be particularly close to the genes it acts on, and need not be located on the same chromosome.
In eukaryotic cells the structure of the chromatin complex of DNA is folded in a way that functionally mimics the supercoiled state characteristic of prokaryotic DNA, so that although the enhancer DNA is far from the gene in regard to the number of nucleotides, it is geometrically close to the promoter and gene. This allows it to interact with the general transcription factors and RNA polymerase II. An enhancer may be located upstream or downstream of the gene that it regulates.
Furthermore, an enhancer does not need to be located near to the transcription initiation site to affect the transcription of a gene, as some have been found to bind several hundred thousand base pairs upstream or downstream of the start site. Enhancers do not act on the promoter region itself, but are bound by activator proteins. These activator proteins interact with the mediator complex, which recruits polymerase II and the general transcription factors which then begin transcribing the genes. Enhancers can also be found within introns. An enhancer's orientation may even be reversed without affecting its function. Additionally, an enhancer may be excised and inserted elsewhere in the chromosome, and still affect gene transcription. That is the reason that intron polymorphisms are checked though they are not translated.
Corepressor
editA corepressor is a protein that decreases gene expression by binding to a transcription factor which contains a DNA binding domain. The corepressor is unable to bind DNA by itself. The corepressor can repress transcriptional initiation by recruiting histone deacetylases which catalyze the removal of acetyl groups from lysine residues. This increases the positive charge on histones which strengthens in the interaction between the histones and DNA, making the latter less accessible to transcription.
Riboswitch
editIn molecular biology, a riboswitch is a part of an mRNA molecule that can directly bind a small target molecule, and whose binding of the target affects the gene's activity. Thus, an mRNA that contains a riboswitch is directly involved in regulating its own activity, in response to the concentrations of its target molecule. The discovery that modern organisms use RNA to bind small molecules, and discriminate against closely related analogs, significantly expanded the known natural capabilities of RNA beyond its ability to code for proteins or to bind other RNA or protein macromolecules. The original definition of the term "riboswitch" specified that they directly sense small-molecule metabolite concentrations. Although this definition remains in common use, some biologists have used a broader definition that includes other cis-regulatory RNAs. However, this article will discuss only metabolite-binding riboswitches. Most known riboswitches occur in bacteria, but functional riboswitches of one type (the TPP riboswitch) have been discovered in plants and certain fungi. TPP riboswitches have also been predicted in archaea, but have not been experimentally tested.[6]
Lac operon
editThe lac operon is an operon required for the transport and metabolism of lactose in Escherichia coli and some other enteric bacteria. It consists of three adjacent structural genes, lacZ, lacY and lacA. The lac operon is regulated by several factors including the availability of glucose and of lactose. Gene regulation of the lac operon was the first complex genetic regulatory mechanism to be elucidated and is one of the foremost examples of prokaryotic gene regulation.
In its natural environment, the lac operon allows for the effective digestion of lactose. The cell can use lactose as an energy source by producing the enzyme β-galactosidase to digest that lactose into glucose and galactose. However, it would be inefficient to produce enzymes when there is no lactose available, or if there is a more readily-available energy source available such as glucose. The lac operon uses a two-part control mechanism to ensure that the cell expends energy producing β-galactosidase, β-galactoside permease and thiogalactoside transacetylase (also known as galactoside O-acetyltransferase) only when necessary. It achieves this with the lac repressor, which halts production in the absence of lactose, and the Catabolite activator protein (CAP), which assists in production in the absence of glucose. This dual control mechanism causes the sequential utilization of glucose and lactose in two distinct growth phases, known as diauxie. Similar diauxic growth patterns have been observed in bacterial growth on mixtures of other sugars as well, such as mixtures of glucose and xylose, or of glucose and arabinose, etc. The genetic control mechanisms underlying such diauxic growth patterns are known as xyl operon and ara operon, etc.[7] The lac operon consists of three structural genes, and a promoter, a terminator, regulator, and an operator.
The three structural genes are: lacZ, lacY, and lacA.
lacZ encodes β-galactosidase (LacZ), an intracellular enzyme that cleaves the disaccharide lactose into glucose and galactose.
lacY encodes β-galactoside permease (LacY), a membrane-bound transport protein that pumps lactose into the cell.
lacA encodes β-galactoside transacetylase (LacA), an enzyme that transfers an acetyl group from acetyl-CoA to β-galactosides.
Only lacZ and lacY appear to be necessary for lactose catabolism.
Lac repressor (LacI)
editThe lac repressor was first isolated by Walter Gilbert and Benno Müller-Hill in 1966. They were able to show, in vitro, that the protein bound to DNA containing the lac operon, and released the DNA when IPTG was added. (IPTG is an allolactose analog.) They were also able to isolate the portion of DNA bound by the protein by using the enzyme deoxyribonuclease, which breaks down DNA. After treatment of the repressor-DNA complex, some DNA remained, suggesting that it had been masked by the repressor. This was later confirmed. These experiments were important, as they confirmed the mechanism of the lac operon, earlier proposed by Jacques Monod and Francois Jacob. The structure of the lac repressor protein consists of three distinct regions:
a core region (which binds allolactose) a tetramerization region (which joins four monomers in an alpha-helix bundle) a DNA-binding region (in which two LacI proteins bind a single operator site) The lac repressor occurs as a tetramer (four identical subunits bound together). This can be viewed as two dimers, with each dimer being able to bind to a single lac operator. The two subunits each bind to a slightly separated (major groove) region of the operator. The promoter is slightly covered by the lac repressor so RNAP cannot bind to and transcribe the operon. The DNA binding region consists of a helix-turn-helix structural motif. Interactive, rotating 3D views of the repressor structure, some bound to DNA, including morphs of how it bends the DNA double helix, are available at Lac Repressor in Proteopedia. The lac repressor (LacI) operates by binding to the major groove of the operator region of the lac operon. This blocks RNA polymerase from binding, and so prevents transcription of the mRNA coding for the Lac proteins. When lactose is present, allolactose binds to the lac repressor, causing an allosteric change in its shape. In its changed state, the lac repressor is unable to bind to its cognate operator.
The lac gene and its derivatives are amenable to use as a reporter gene in a number of bacterial-based selection techniques such as two hybrid analysis, in which the successful binding of a transcriptional activator to a specific promoter sequence must be determined. In LB plates containing X-gal, the colour change from white colonies to a shade of blue corresponds to about 20-100 β-galactosidase units, while tetrazolium lactose and MacConkey lactose media have a range of 100-1000 units, being most sensitive in the high and low parts of this range respectively. Since MacConkey lactose and tetrazolium lactose media both rely on the products of lactose breakdown, they require the presence of both lacZ and lacY genes. The many lac fusion techniques which include only the lacZ gene are thus suited to the X-gal plates or ONPG liquid broths.[8]
Trp operon
editTrp operon is an operon - a group of genes that are used, or transcribed, together - that codes for the components for production of tryptophan. The Trp operon is present in many bacteria, but was first characterized in Escherichia coli. It is regulated so that when tryptophan is present in the environment, it is not used. It was an important experimental system for learning about gene regulation, and is commonly used to teach gene regulation.
Discovered in 1953 by Jacques Monod and colleagues, the trp operon in E. coli was the first repressible operon to be discovered. While the lac operon can be activated by a chemical (allolactose), the tryptophan (Trp) operon is inhibited by a chemical (tryptophan). This operon contains five structural genes: trp E, trp D, trp C, trp B, and trp A, which encodes tryptophan synthetase. It also contains a promoter which binds to RNA polymerase and an operator which blocks transcription when bound to the protein synthesized by the repressor gene (trp R) that binds to the operator. In the lac operon, allolactose binds to the repressor protein, allowing gene transcription, while in the trp operon, tryptophan binds to the repressor protein effectively blocking gene transcription. In both situations, repression is that of RNA polymerase transcribing the genes in the operon. Also unlike the lac operon, the trp operon contains a leader peptide and an attenuator sequence which allows for graded regulation.
It is an example of negative regulation of gene expression. Within the operon's regulatory sequence, the operator is blocked by the repressor protein in the presence of tryptophan (thereby preventing transcription) and is liberated in tryptophan's absence (thereby allowing transcription). The process of attenuation complements this regulatory action.[9]
Arabinose operon
editThe L-arabinose operon of the model bacterium Escherichia coli has been a focus for research in molecular biology for over 40 years, and has been investigated extensively at the genetic, biochemical, physiological, and biophysical levels. It is controlled by a dual positive and negative system. There are 3 structural genes: araB, araA, and araD. They encode the metabolic enzymes for breaking down the monosaccharide sugar arabinose into D-xylulose-5-phosphate, which is then metabolised via the pentose phosphate pathway. The initiator region, containing an operator site as well as a promoter, is called araI (the last letter of araI is an uppercase letter " i "). Near this site lies the araC gene, which encodes a repressor protein. The AraC protein binds to initiator region araI.
Housekeeping gene
editA housekeeping gene is typically a constitutive gene that is required for the maintenance of basic cellular function, and are found in all human cells. Although some housekeeping genes are expressed at relatively constant levels(such as HSP90 and Beta-actin), other housekeeping genes may vary depending on experimental conditions. The origin of the term "housekeeping gene" remains obscure. Literature from 1976 used the term to describe specifically tRNA and rRNA. Interpreting gene expression data can be problematic, with most human genes registering 5-10 copies per cell (possibly representing error). Housekeeping genes are expressed in at least 25 copies per cell and sometimes number in the thousands.
Regulation of gene expression
editRegulation of gene expression refers to the control of the amount and timing of appearance of the functional product of a gene. Control of expression is vital to allow a cell to produce the gene products it needs when it needs them; in turn this gives cells the flexibility to adapt to a variable environment, external signals, damage to the cell, etc. Some simple examples of where gene expression is important are:
Control of Insulin expression so it gives a signal for blood glucose regulation
X chromosome inactivation in female mammals to prevent an "overdose" of the genes it contains.
Cyclin expression levels control progression through the eukaryotic cell cycle
More generally gene regulation gives the cell control over all structure and function, and is the basis for cellular differentiation, morphogenesis and the versatility and adaptability of any organism. Any step of gene expression may be modulated, from the DNA-RNA transcription step to post-translational modification of a protein. The stability of the final gene product, whether it is RNA or protein, also contributes to the expression level of the gene - an unstable product results in a low expression level. In general gene expression is regulated through changes in the number and type of interactions between molecules that collectively influence transcription of DNA and translation of RNA. Numerous terms are used to describe types of genes depending on how they are regulated, these include: A constitutive gene is a gene that is transcribed continually compared to a facultative gene which is only transcribed when needed. A housekeeping gene is typically a constitutive gene that is transcribed at a relatively constant level. The housekeeping gene's products are typically needed for maintenance of the cell. It is generally assumed that their expression is unaffected by experimental conditions. Examples include actin, GAPDH and ubiquitin. A facultative gene is a gene which is only transcribed when needed compared to a constitutive gene. An inducible gene is a gene whose expression is either responsive to environmental change or dependent on the position in the cell cycle.[10]
Transcriptional regulation Regulation of transcription can be broken down into three main routes of influence; genetic (direct interaction of a control factor with the gene), modulation (interaction of a control factor with the transcription machinery) and epigenetic (non-sequence changes in DNA structure which influence transcription).
The lambda repressor transcription factor (green) binds as a dimer to major groove of DNA target (red and blue) and disables initiation of transcription. From PDB 1LMB. Direct interaction with DNA is the simplest and the most direct method by which a protein can change transcription levels. Genes often have several protein binding sites around the coding region with the specific function of regulating transcription. There are many classes of regulatory DNA binding sites known as enhancers, insulators, repressors and silencers. The mechanisms for regulating transcription are very varied, from blocking key binding sites on the DNA for RNA polymerase to acting as an activator and promoting transcription by assisting RNA polymerase binding. The activity of transcription factors is further modulated by intracellular signals causing protein post-translational modification including phosphorylated, acetylated, or glycosylated. These changes influence a transcription factor's ability to bind, directly or indirectly, to promoter DNA, to recruit RNA polymerase, or to favor elongation of a newly synthetized RNA molecule. The nuclear membrane in eukaryotes allows further regulation of transcription factors by the duration of their presence in the nucleus which is regulated by reversible changes in their structure and by binding of other proteins. Environmental stimuli or endocrine signals may cause modification of regulatory proteins eliciting cascades of intracellular signals, which result in regulation of gene expression. More recently it has become apparent that there is a huge influence of non-DNA-sequence specific effects on translation. These effects are referred to as epigenetic and involve the higher order structure of DNA, non-sequence specific DNA binding proteins and chemical modification of DNA. In general epigenetic effects alter the accessibility of DNA to proteins and so modulate transcription.
In eukaryotes, DNA is organized in form of nucleosomes. Note how the DNA (blue and green) is tightly wrapped around the protein core made of histone octamer (ribbon coils), restricting access to the DNA. From PDB 1KX5. DNA methylation is a widespread mechanism for epigenetic influence on gene expression and is seen in bacteria and eukaryotes and has roles in heritable transcription silencing and transcription regulation. In eukaryotes the structure of chromatin, controlled by the histone code, regulates access to DNA with significant impacts on the expression of genes in euchromatin and heterochromatin areas.
Post-transcriptional regulation
In eukaryotes, where export of RNA is required before translation is possible, nuclear export is thought to provide additional control over gene expression. All transport in and out of the nucleus is via the nuclear pore and transport is controlled by a wide range of importin and exportin proteins. Expression of a gene coding for a protein is only possible if the messenger RNA carrying the code survives long enough to be translated. In a typical cell an RNA molecule is only stable if specifically protected from degradation. RNA degradation has particular importance in regulation of expression in eukaryotic cells where mRNA has to travel significant distances before being translated. In eukaryotes RNA is stabilised by certain post-transcriptional modifications, particularly the 5' cap and poly-adenylated tail. Intentional degradation of mRNA is used not just as a defence mechanism from foreign RNA (normally from viruses) but also as a route of mRNA destabilisation. If an mRNA molecule has a complementary sequence to a small interfering RNA then it is targeted for destruction via the RNA interference pathway.
Translational regulation
Neomycin is an example of a small molecule which reduces expression of all protein genes inevitably leading to cell death, thus acts as an antibiotic.
Direct regulation of translation is less prevalent than control of transcription or mRNA stability but is occasionally used. Inhibition of protein translation is a major target for toxins and antibiotics in order to kill a cell by overriding its normal gene expression control. Protein synthesis inhibitors include the antibiotic neomycin and the toxin ricin. Protein degradation
Once protein synthesis is complete the level of expression of that protein can be reduced by protein degradation. There are major protein degradation pathways in all prokaryotes and eukaryotes of which the proteasome is a common component. An unneeded or damaged protein is often labelled for degradation by addition of ubiquitin.
Tools for studying gene expression
editVector
editPlasmids used in genetic engineering are called vectors. Plasmids serve as important tools in genetics and biotechnology labs, where they are commonly used to multiply (make many copies of) or express particular genes. Many plasmids are commercially available for such uses. The gene to be replicated is inserted into copies of a plasmid containing genes that make cells resistant to particular antibiotics and a multiple cloning site (MCS, or polylinker), which is a short region containing several commonly used restriction sites allowing the easy insertion of DNA fragments at this location. Next, the plasmids are inserted into bacteria by a process called transformation. Then, the bacteria are exposed to the particular antibiotics. Only bacteria which take up copies of the plasmid survive, since the plasmid makes them resistant. In particular, the protecting genes are expressed (used to make a protein) and the expressed protein breaks down the antibiotics. In this way the antibiotics act as a filter to select only the modified bacteria. Now these bacteria can be grown in large amounts, harvested and lysed (often using the alkaline lysis method) to isolate the plasmid of interest. Another major use of plasmids is to make large amounts of proteins. In this case, researchers grow bacteria containing a plasmid harboring the gene of interest. Just as the bacteria produces proteins to confer its antibiotic resistance, it can also be induced to produce large amounts of proteins from the inserted gene. This is a cheap and easy way of mass-producing a gene or the protein it then codes for, for example, insulin or even antibiotics. However, a plasmid can only contain inserts of about 1–10 kbp. To clone longer lengths of DNA, lambda phage with lysogeny genes deleted, cosmids, bacterial artificial chromosomes or yeast artificial chromosomes could be used.[11]
Modern vectors may encompass additional features besides the transgene insert and a backbone: Promoter: Necessary component for all vectors: used to drive transcription of the vector's transgene.
Genetic markers: Genetic markers for viral vectors allow for confirmation that the vector has integrated with the host genomic DNA.
Antibiotic resistance: Vectors with antibiotic-resistance open reading frames allow for survival of cells that have taken up the vector in growth media containing antibiotics through antibiotic selection.
Epitope: Vector contains a sequence for a specific epitope that is incorporated into the expressed protein. Allows for antibody identification of cells expressing the target protein.
β-galactosidase: Some vectors contain a sequence for β-galactosidase, an enzyme that digests galactose, within which a multiple cloning site, the region in which a gene may be inserted, is located. An insert successfully ligated into the vector will disrupt the β-galactosidase gene and disable galactose digestion. Cells containing vector with an insert may be identified using blue/white selection by growing cells in media containing an analogue of galactose (X-gal). Cells expressing β-galactosidase (therefore doesn't contain an insert) appear as blue colonies. White colonies would be selected as those that may contain an insert. Other proteins which may function similarly as a reporter include green fluorescent protein and luciferase.
Targeting sequence: Expression vectors may include encoding for a targeting sequence in the finished protein that directs the expressed protein to a specific organelle in the cell or specific location such as the periplasmic space of bacteria.
Protein purification tags: Some expression vectors include proteins or peptide sequences that allows for easier purification of the expressed protein. Examples include polyhistidine-tag, glutathione-S-transferase, and maltose binding protein. Some of these tags may also allow for increased solubility of the target protein. The target protein is fused to the protein tag, but a protease cleavage site positioned in the polypeptide linker region between the protein and the tag allows the tag to be removed later. Cosmids Cosmids are predominantly plasmids with a bacterial oriV, an antibiotic selection marker and a cloning site, but they carry one, or more recently two cos sites derived from bacteriophage lambda. Depending on the particular aim of the experiment broad host range cosmids, shuttle cosmids or 'mammalian' cosmids (linked to SV40 oriV and mammalian selection markers) are available. The loading capacity of cosmids varies depending on the size of the vector itself but usually lies around 40–45 kb. The cloning procedure involves the generation of two vector arms which are then joined to the foreign DNA. Selection against wildtype cosmid DNA is simply done via size exclusion. Cosmids, however, always form colonies and not plaques. Also the clone density is much lower with around 105 - 106 CFU per µg of ligated DNA. After the construction of recombinant lambda or cosmid libraries the total DNA is transferred into an appropriate E.coli host via a technique called in vitro packaging. The necessary packaging extracts are derived from E.coli cI857 lysogens (red- gam- Sam and Dam (head assembly) and Eam (tail assembly) respectively). These extracts will recognize and package the recombinant molecules in vitro, generating either mature phage particles (lambda-based vectors) or recombinant plasmids contained in phage shells (cosmids). These differences are reflected in the different infection frequencies seen in favour of lambda-replacement vectors. This compensates for their slightly lower loading capacity. Phage library are also stored and screened easier than cosmid (colonies!) libraries. Target DNA: the genomic DNA to be cloned has to be cut into the appropriate size range of restriction fragments. This is usually done by partial restriction followed by either size fractionation or dephosphorylation (using calf-intestine phosphatase) to avoid chromosome scrambling, i.e. the ligation of physically unlinked fragments. Fosmids Fosmids are similar to cosmids but are based on the bacterial F-plasmid. The cloning vector is limited, as a host (usually E. coli) can only contain one fosmid molecule. Fosmids are 40 kb of random genomic DNA. Fosmid library is prepared from a genome of the target organism and cloned into a fosmid vector. Low copy number offers higher stability than comparable high copy number cosmids. Fosmid system may be useful for constructing stable libraries from complex genomes. Fosmid clones were used to help assess the accuracy of the Public Human Genome Sequence.
Bacterial artificial chromosome (BAC) A bacterial artificial chromosome (BAC) is a DNA construct, based on a functional fertility plasmid (or F-plasmid), used for transforming and cloning in bacteria, usually E. coli. F-plasmids play a crucial role because they contain partition genes that promote the even distribution of plasmids after bacterial cell division. The bacterial artificial chromosome's usual insert size is 150-350 kbp, but can be greater than 700 kbp. A similar cloning vector called a PAC has also been produced from the bacterial P1-plasmid. BACs are often used to sequence the genome of organisms in genome projects, for example the Human Genome Project. A short piece of the organism's DNA is amplified as an insert in BACs, and then sequenced. Finally, the sequenced parts are rearranged in silico, resulting in the genomic sequence of the organism.
Yeast artificial chromosome (YAC) A yeast artificial chromosome (YAC) is a vector used to clone DNA fragments larger than 100 kb and up to 3000 kb. YACs are useful for the physical mapping of complex genomes and for the cloning of large genes. First described in 1983 by Murray and Szostak, a YAC is an artificially constructed chromosome and contains the telomeric, centromeric, and replication origin sequences needed for replication and preservation in yeast cells. A YAC is built using an initial circular plasmid, which is typically broken into two linear molecules using restriction enzymes; DNA ligase is then used to ligate a sequence or gene of interest between the two linear molecules, forming a single large linear piece of DNA.[citation needed] Yeast expression vectors, such as YACs, YIps (yeast integrating plasmids), and YEps (yeast episomal plasmids), have an advantage over bacterial artificial chromosomes (BACs) in that they can be used to express eukaryotic proteins that require posttranslational modification. However, YACs have been found to be less stable than BACs, producing chimeric effects.
Types of viral vectors
Retroviruses
Retroviruses are one of the mainstays of current gene therapy approaches. The recombinant retroviruses such as the Moloney murine leukemia virus have the ability to integrate into the host genome in a stable fashion. They contain a reverse transcriptase that allows integration into the host genome. They have been used in a number of FDA-approved clinical trials such as the SCID-X1 trial. Retroviral vectors can either be replication-competent or replication-defective. Replication-defective vectors are the most common choice in studies because the viruses have had the coding regions for the genes necessary for additional rounds of virion replication and packaging replaced with other genes, or deleted. These virus are capable of infecting their target cells and delivering their viral payload, but then fail to continue the typical lytic pathway that leads to cell lysis and death. Conversely, replication-competent viral vectors contain all necessary genes for virion synthesis, and continue to propagate themselves once infection occurs. Because the viral genome for these vectors is much lengthier, the length of the actual inserted gene of interest is limited compared to the possible length of the insert for replication-defective vectors. Depending on the viral vector, the typical maximum length of an allowable DNA insert in a replication-defective viral vector is usually about 8–10 kB. While this limits the introduction of many genomic sequences, most cDNA sequences can still be accommodated. The primary drawback to use of retroviruses such as the Moloney retrovirus involves the requirement for cells to be actively dividing for transduction. As a result, cells such as neurons are very resistant to infection and transduction by retroviruses. There is concern that insertional mutagenesis due to integration into the host genome might lead to cancer or leukemia.
Lentiviruses
Lentiviruses are a subclass of Retroviruses. They have recently been adapted as gene delivery vehicles (vectors) thanks to their ability to integrate into the genome of non-dividing cells, which is the unique feature of Lentiviruses as other Retroviruses can infect only dividing cells. The viral genome in the form of RNA is reverse-transcribed when the virus enters the cell to produce DNA, which is then inserted into the genome at a random position by the viral integrase enzyme. The vector, now called a provirus, remains in the genome and is passed on to the progeny of the cell when it divides. The site of integration is unpredictable, which can pose a problem. The provirus can disturb the function of cellular genes and lead to activation of oncogenes promoting the development of cancer, which raises concerns for possible applications of lentiviruses in gene therapy. However, studies have shown that lentivirus vectors have a lower tendency to integrate in places that potentially cause cancer than gamma-retroviral vectors. More specifically, one study found that lentiviral vectors did not cause either an increase in tumor incidence or an earlier onset of tumors in a mouse strain with a much higher incidence of tumors. Moreover, clinical trials that utilized lentiviral vectors to deliver gene therapy for the treatment of HIV experienced no increase in mutagenic or oncologic events. For safety reasons lentiviral vectors never carry the genes required for their replication. To produce a lentivirus, several plasmids are transfected into a so-called packaging cell line, commonly HEK 293. One or more plasmids, generally referred to as packaging plasmids, encode the virion proteins, such as the capsid and the reverse transcriptase. Another plasmid contains the genetic material to be delivered by the vector. It is transcribed to produce the single-stranded RNA viral genome and is marked by the presence of the ψ (psi) sequence. This sequence is used to package the genome into the virion.
Adenoviruses
As opposed to lentiviruses, adenoviral DNA does not integrate into the genome and is not replicated during cell division. This limits their use in basic research, although adenoviral vectors are occasionally used in in vitro experiments. Their primary applications are in gene therapy and vaccination. Since humans commonly come in contact with adenoviruses, which cause respiratory, gastrointestinal and eye infections, they trigger a rapid immune response with potentially dangerous consequences. To overcome this problem scientists are currently investigating adenoviruses to which humans do not have immunity.
Adeno-associated viruses
Adeno-associated virus (AAV) is a small virus that infects humans and some other primate species. AAV is not currently known to cause disease and consequently the virus causes a very mild immune response. AAV can infect both dividing and non-dividing cells and may incorporate its genome into that of the host cell. These features make AAV a very attractive candidate for creating viral vectors for gene therapy.
PCR
editPCR is used to amplify a specific region of a DNA strand (the DNA target). Most PCR methods typically amplify DNA fragments of up to ~10 kilo base pairs (kb), although some techniques allow for amplification of fragments up to 40 kb in size. A basic PCR set up requires several components and reagents.These components include:
DNA template that contains the DNA region (target) to be amplified.
Two primers that are complementary to the 3' (three prime) ends of each of the sense and anti-sense strand of the DNA target. Taq polymerase or another DNA polymerase with a temperature optimum at around 70 °C. Deoxynucleotide triphosphates (dNTPs), the building-blocks from which the DNA polymerase synthesizes a new DNA strand. Buffer solution, providing a suitable chemical environment for optimum activity and stability of the DNA polymerase. Divalent cations, magnesium or manganese ions; generally Mg2+ is used, but Mn2+ can be utilized for PCR-mediated DNA mutagenesis, as higher Mn2+ concentration increases the error rate during DNA synthesis Monovalent cation potassium ions. The PCR is commonly carried out in a reaction volume of 10–200 μl in small reaction tubes (0.2–0.5 ml volumes) in a thermal cycler. The thermal cycler heats and cools the reaction tubes to achieve the temperatures required at each step of the reaction (see below). Many modern thermal cyclers make use of the Peltier effect, which permits both heating and cooling of the block holding the PCR tubes simply by reversing the electric current. Thin-walled reaction tubes permit favorable thermal conductivity to allow for rapid thermal equilibration. Most thermal cyclers have heated lids to prevent condensation at the top of the reaction tube. Older thermocyclers lacking a heated lid require a layer of oil on top of the reaction mixture or a ball of wax inside the tube.[12]
Procedure
Figure 1: Schematic drawing of the PCR cycle. (1) Denaturing at 94–96 °C. (2) Annealing at ~65 °C (3) Elongation at 72 °C. Four cycles are shown here. The blue lines represent the DNA template to which primers (red arrows) anneal that are extended by the DNA polymerase (light green circles), to give shorter DNA products (green lines), which themselves are used as templates as PCR progresses. Typically, PCR consists of a series of 20-40 repeated temperature changes, called cycles, with each cycle commonly consisting of 2-3 discrete temperature steps, usually three . The cycling is often preceded by a single temperature step (called hold) at a high temperature (>90 °C), and followed by one hold at the end for final product extension or brief storage. The temperatures used and the length of time they are applied in each cycle depend on a variety of parameters. These include the enzyme used for DNA synthesis, the concentration of divalent ions and dNTPs in the reaction, and the melting temperature (Tm) of the primers.Initialization step: This step consists of heating the reaction to a temperature of 94–96 °C (or 98 °C if extremely thermostable polymerases are used), which is held for 1–9 minutes. It is only required for DNA polymerases that require heat activation by hot-start PCR. Denaturation step: This step is the first regular cycling event and consists of heating the reaction to 94–98 °C for 20–30 seconds. It causes DNA melting of the DNA template by disrupting the hydrogen bonds between complementary bases, yielding single-stranded DNA molecules. Annealing step: The reaction temperature is lowered to 50–65 °C for 20–40 seconds allowing annealing of the primers to the single-stranded DNA template. Typically the annealing temperature is about 3-5 degrees Celsius below the Tm of the primers used. Stable DNA-DNA hydrogen bonds are only formed when the primer sequence very closely matches the template sequence. The polymerase binds to the primer-template hybrid and begins DNA synthesis. Extension/elongation step: The temperature at this step depends on the DNA polymerase used; Taq polymerase has its optimum activity temperature at 75–80 °C, and commonly a temperature of 72 °C is used with this enzyme. At this step the DNA polymerase synthesizes a new DNA strand complementary to the DNA template strand by adding dNTPs that are complementary to the template in 5' to 3' direction, condensing the 5'-phosphate group of the dNTPs with the 3'-hydroxyl group at the end of the nascent (extending) DNA strand. The extension time depends both on the DNA polymerase used and on the length of the DNA fragment to be amplified. As a rule-of-thumb, at its optimum temperature, the DNA polymerase will polymerize a thousand bases per minute. Under optimum conditions, i.e., if there are no limitations due to limiting substrates or reagents, at each extension step, the amount of DNA target is doubled, leading to exponential (geometric) amplification of the specific DNA fragment. Final elongation: This single step is occasionally performed at a temperature of 70–74 °C for 5–15 minutes after the last PCR cycle to ensure that any remaining single-stranded DNA is fully extended.[13] Final hold: This step at 4–15 °C for an indefinite time may be employed for short-term storage of the reaction.
To check whether the PCR generated the anticipated DNA fragment (also sometimes referred to as the amplimer or amplicon), agarose gel electrophoresis is employed for size separation of the PCR products. The size(s) of PCR products is determined by comparison with a DNA ladder (a molecular weight marker), which contains DNA fragments of known size, run on the gel alongside the PCR products.
Restriction enzymes
editRestriction enzymes recognize a specific sequence of nucleotides and produce a double-stranded cut in the DNA. While recognition sequences vary between 4 and 8 nucleotides, many of them are palindromic, which correspond to nitrogenous base sequences that read the same backwards and forwards.[14] In theory, there are two types of palindromic sequences that can be possible in DNA. The mirror-like palindrome is similar to those found in ordinary text, in which a sequence reads the same forward and backwards on the same DNA strand (i.e., single stranded) as in GTAATG. The inverted repeat palindrome is also a sequence that reads the same forward and backwards, but the forward and backward sequences are found in complementary DNA strands (i.e., double stranded) as in GTATAC (Notice that GTATAC is complementary to CATATG).[15] The inverted repeat is more common and has greater biological importance than the mirror-like.
5'-GTATAC-3' :::::: 3'-CATATG-5' |
| A palindromic recognition site reads the same on the reverse strand as it does on the forward strand |
EcoRI digestion produces "sticky" ends,
whereas SmaI restriction enzyme cleavage produces "blunt" ends
Recognition sequences in DNA differ for each restriction enzyme, producing differences in the length, sequence and strand orientation (5' end or the 3' end) of a sticky-end "overhang" of an enzyme restriction.[16]
Different restriction enzymes that recognize the same sequence are known as neoschizomers. These often cleave in a different locales of the sequence; however, different enzymes that recognize and cleave in the same location are known as an isoschizomer.
Classification of Restriction Enzymes
Restriction endonucleases are categorized into three or four general groups (Types I, II and III) based on their composition and enzyme cofactor requirements, the nature of their target sequence, and the position of their DNA cleavage site relative to the target sequence. There are four classes of restriction endonucleases: types I, II,III and IV. All types of enzymes recognise specific short DNA sequences and carry out the endonucleolytic cleavage of DNA to give specific double-stranded fragments with terminal 5'-phosphates. They differ in their recognition sequence, subunit composition, cleavage position, and cofactor requirements, as summarised below:
Type I enzymes (EC 3.1.21.3) cleave at sites remote from recognition site; require both ATP and S-adenosyl-L-methionine to function; multifunctional protein with both restriction and methylase (EC 2.1.1.72) activities.
Type II enzymes (EC 3.1.21.4) cleave within or at short specific distances from recognition site; most require magnesium; single function (restriction) enzymes independent of methylase.
Type III enzymes (EC 3.1.21.5) cleave at sites a short distance from recognition site; require ATP (but doesn't hydrolyse it); S-adenosyl-L-methionine stimulates reaction but is not required; exist as part of a complex with a modification methylase (EC 2.1.1.72). Type IV enzymes target methylated DNA.
Examples of restriction enzymes include:[17]
| Enzyme | Source | Recognition Sequence | Cut |
|---|---|---|---|
| EcoRI | Escherichia coli |
5'GAATTC 3'CTTAAG |
5'---G AATTC---3' 3'---CTTAA G---5' |
| EcoRII | Escherichia coli |
5'CCWGG 3'GGWCC |
5'--- CCWGG---3' 3'---GGWCC ---5' |
| BamHI | Bacillus amyloliquefaciens |
5'GGATCC 3'CCTAGG |
5'---G GATCC---3' 3'---CCTAG G---5' |
| HindIII | Haemophilus influenzae |
5'AAGCTT 3'TTCGAA |
5'---A AGCTT---3' 3'---TTCGA A---5' |
| TaqI | Thermus aquaticus |
5'TCGA 3'AGCT |
5'---T CGA---3' 3'---AGC T---5' |
| NotI | Nocardia otitidis |
5'GCGGCCGC 3'CGCCGGCG |
5'---GC GGCCGC---3' 3'---CGCCGG CG---5' |
| HinfI | Haemophilus influenzae |
5'GANTCA 3'CTNAGT |
5'---G ANTC---3' 3'---CTNA G---5' |
| Sau3A | Staphylococcus aureus |
5'GATC 3'CTAG |
5'--- GATC---3' 3'---CTAG ---5' |
| PovII* | Proteus vulgaris |
5'CAGCTG 3'GTCGAC |
5'---CAG CTG---3' 3'---GTC GAC---5' |
| SmaI* | Serratia marcescens |
5'CCCGGG 3'GGGCCC |
5'---CCC GGG---3' 3'---GGG CCC---5' |
| HaeIII* | Haemophilus aegyptius |
5'GGCC 3'CCGG |
5'---GG CC---3' 3'---CC GG---5' |
| HgaI[18] | Haemophilus gallinarum |
5'GACGC 3'CTGCG |
5'---NN NN---3' 3'---NN NN---5' |
| AluI* | Arthrobacter luteus |
5'AGCT 3'TCGA |
5'---AG CT---3' 3'---TC GA---5' |
| EcoRV* | Escherichia coli |
5'GATATC 3'CTATAG |
5'---GAT ATC---3' 3'---CTA TAG---5' |
| EcoP15I | Escherichia coli |
5'CAGCAGN25NN 3'GTCGTCN25NN |
5'---CAGCAGN25NN ---3' 3'---GTCGTCN25 NN---5' |
| KpnI | Klebsiella pneumoniae |
5'GGTACC 3'CCATGG |
5'---GGTAC C---3' 3'---C CATGG---5' |
| PstI | Providencia stuartii |
5'CTGCAG 3'GACGTC |
5'---CTGCA G---3' 3'---G ACGTC---5' |
| SacI | Streptomyces achromogenes |
5'GAGCTC 3'CTCGAG |
5'---GAGCT C---3' 3'---C TCGAG---5' |
| SalI | Streptomyces albus |
5'GTCGAC 3'CAGCTG |
5'---G TCGAC---3' 3'---CAGCT G---5' |
| ScaI | Streptomyces caespitosus |
5'AGTACT 3'TCATGA |
5'---AGT ACT---3' 3'---TCA TGA---5' |
| SpeI | Sphaerotilus natans |
5'ACTAGT 3'TGATCA |
5'---A CTAGT---3' 3'---TGATC A---5' |
| SphI | Streptomyces phaeochromogenes |
5'GCATGC 3'CGTACG |
5'---GCATG C---3' 3'---C GTACG---5' |
| StuI[19][20] | Streptomyces tubercidicus |
5'AGGCCT 3'TCCGGA |
5'---AGG CCT---3' 3'---TCC GGA---5' |
| XbaI | Xanthomonas badrii |
5'TCTAGA 3'AGATCT |
5'---T CTAGA---3' 3'---AGATC T---5' |
Key:
* = blunt ends
N = C or G or T or A
W = A or T
Cloning of gene and its expression
editIn molecular biology Cloning refers to the procedure of isolating a defined DNA sequence and obtaining multiple copies of it in vitro. Cloning is frequently employed to amplify DNA fragments containing genes, but it can be used to amplify any DNA sequence such as promoters, non-coding sequences, chemically synthesised oligonucleotides and randomly fragmented DNA. Cloning is used in a wide array of biological experiments and technological applications such as large scale protein production and expression of gene in cell lines like HeLA cells.
In essence, in order to amplify any DNA sequence in vivo and in vitro, the sequence in question must be linked to primary sequence elements capable of directing the replication and propagation of themselves and the linked sequence in the desired target host. The required sequence elements differ according to host, but invariably include an origin of replication, and a selectable marker. In practice, however, a number of other features are desired and a variety of specialized cloning vectors exist that allow protein expression, tagging, single stranded RNA and DNA production and a host of other manipulations that are useful in downstream applications.
Recombinase-based cloning A novel procedure of cloning or subcloning of any DNA fragment by inserting the special DNA fragment of interest into a special area of target DNA through interchange of the relevant DNA fragments.[21]
This is a one-step reaction: simple, efficient, facilitating high throughput or automatic cloning and/or subcloning.[22]
Restriction/ligation cloning In the classical restriction and ligation cloning protocols, cloning of any DNA fragment essentially involves four steps: DNA fragmentation with restriction endonucleases, ligation of DNA fragments to a vector, transfection, and screening/selection. Although these steps are invariable among cloning procedures a number of alternative routes can be selected at various points depending on the particular application; these are summarized as a ‘cloning strategy’.
Isolation of insert Initially, the DNA fragment to be cloned needs to be isolated. Preparation of DNA fragments for cloning can be accomplished in a number of alternative ways. Insert preparation is frequently achieved by means of polymerase chain reaction, but it may also be accomplished by restriction enzyme digestion, DNA sonication and fractionation by agarose gel electrophoresis. Chemically synthesized oligonucleotides can also be used if the target sequence size does not exceed the limit of chemical synthesis. Isolation of insert can be done by using shotgun cloning, c-DNA clones, gene machines (artificial chemical synthesis).
Transformation
Following ligation, the ligation product (plasmid) is transformed into bacteria for propagation. The bacteria is then plated on selective agar to select for bacteria that have the plasmid of interest. Individual colonies are picked and tested for the wanted insert. Maxiprep can be done to obtain large quantity of the plasmid containing the inserted gene.
Transfection Following ligation, a portion of the ligation reaction, including vector with insert in the desired orientation is transfected into cells. A number of alternative techniques are available, such as chemical sensitization of cells, electroporation and biolistics. Chemical sensitization of cells is frequently employed since this does not require specialized equipment and provides relatively high transformation efficiencies. Electroporation is used when extremely high transformation efficiencies are required, as in very inefficient cloning strategies. Biolistics are mainly utilized in plant cell transformations, where the cell wall is a major obstacle in DNA uptake by cells. The bacterial transformation is generally observed by blue white screening.
Selection Finally, the transfected cells are cultured. As the aforementioned procedures are of particularly low efficiency, there is a need to identify the cells that contain the desired insert at the appropriate orientation and isolate these from those not successfully transformed. Modern cloning vectors include selectable markers (most frequently antibiotic resistance markers) that allow only cells in which the vector, but not necessarily the insert, has been transfected to grow. Additionally, the cloning vectors may contain colour selection markers which provide blue/white screening (via α-factor complementation) on X-gal medium. Nevertheless, these selection steps do not absolutely guarantee that the DNA insert is present in the cells. Further investigation of the resulting colonies is required to confirm that cloning was successful. This may be accomplished by means of PCR, restriction fragment analysis and/or DNA sequencing.
Genetic engineering
Genetic engineering is a method of changing the inherited characteristics of an organism in a predetermined way by altering its genetic material. This is often done to enable micro-organisms, such as bacteria or viruses, to synthesize increased yields of compounds, to form entirely new compounds, or to adapt to different environments. Other uses of this technology, which is also called recombinant DNA technology, include gene therapy, which is the supply of a functional gene to a person with a genetic disorder or with other diseases such as acquired immune deficiency syndrome (AIDS) or cancer, and the cloning of whole organisms.
Genetic engineering involves the manipulation of deoxyribonucleic acid, or DNA. Important tools in this process are restriction endonucleases (so-called restriction enzymes) that are produced by various species of bacteria. Restriction enzymes can recognize a particular sequence of the chain of chemical units, called nucleotide bases, which make up the DNA molecule and cut the DNA at that location. Fragments of DNA generated in this way can be joined using other enzymes called ligases. Restriction enzymes and ligases therefore allow the specific cutting and reassembling of portions of DNA. Also important in the manipulation of DNA are so-called vectors, which are pieces of DNA that can self-replicate (produce copies of themselves) independently of the DNA in the host cell in which they are grown. Examples of vectors include plasmids, viruses, and artificial chromosomes. Vectors permit the generation of multiple copies of a particular piece of DNA, making this a useful method for generating sufficient quantities of material with which to work. The process of engineering a DNA fragment into a vector is called “molecular cloning”, because multiple copies of an identical molecule of DNA are produced. Another way of producing many identical copies of a particular (often short, for example, 100-3,000 base pairs) DNA fragment is the polymerase chain reaction. This method is rapid and avoids the need for cloning DNA into a vector.
Reporter gene
editIn molecular biology, a reporter gene is a gene that researchers attach to a regulatory sequence of another gene of interest in cell culture, animals or plants. Certain genes are chosen as reporters because the characteristics they confer on organisms expressing them are easily identified and measured, or because they are selectable markers. Reporter genes are often used as an indication of whether a certain gene has been taken up by or expressed in the cell or organism population.
To introduce a reporter gene into an organism, scientists place the reporter gene and the gene of interest in the same DNA construct to be inserted into the cell or organism. For bacteria or eukaryotic cells in culture, this is usually in the form of a circular DNA molecule called a plasmid. It is important to use a reporter gene that is not natively expressed in the cell or organism under study, since the expression of the reporter is being used as a marker for successful uptake of the gene of interest. Commonly used reporter genes that induce visually identifiable characteristics usually involve fluorescent and luminescent proteins; examples include the gene that encodes jellyfish green fluorescent protein (GFP), which causes cells that express it to glow green under blue light, the enzyme luciferase, which catalyzes a reaction with luciferin to produce light, and the red fluorescent protein from the gene dsRed. Another common reporter in bacteria is the Lac Z gene, which encodes the protein beta-galactosidase. This enzyme causes bacteria expressing the gene to appear blue when grown on a medium that contains the substrate analog X-gal. An example of a selectable-marker reporter in bacteria is the chloramphenicol acetyltransferase (CAT) gene, which confers resistance to the antibiotic chloramphenicol.
Reporter genes can also be used to assay for the expression of the gene of interest, which may produce a protein that has little obvious or immediate effect on the cell culture or organism. In these cases the reporter is directly attached to the gene of interest to create a gene fusion. The two genes are under the same promoter elements and are transcribed into a single messenger RNA molecule. The mRNA is then translated into protein. In these cases it is important that both proteins be able to properly fold into their active conformations and interact with their substrates despite being fused. In building the DNA construct, a segment of DNA coding for a flexible polypeptide linker region is usually included so that the reporter and the gene product will only minimally interfere with one another.[23]
Gene expression and purification in Practice
Protein expression is crucial in biochemistry as it provides the substrate or enzyme required for further analysis. Before large scale protein expression, a small scale expression checked is usually first applied. BL21 Competent E. coli is a commonly used protein expression competent cells. It contains resistant to certain antibiotics, such as Kanamycin; it can undergo modification to express proteins of interest.
In an expression check, desired genome is inoculated and expressed overnight in 5ml appropriate media with corresponding antibiotics. After overnight expression, the media containing desired protein is spun down. After supernatant is removed, the pellet is suspended into appropriate lysis buffer and sonicated. After sonication, the sample is spun down. The soluble fraction and insoluble fractions are taken for gel analysis on an SDS page. The approximate size of the desired protein must be known and calculated ahead. If desire band shows up in the SDS page, large scale protein expression can then move on.
In a large scale protein expression, three liter cultural flasks are commonly used for inoculation and induction. At the beginning, starter culture of gene of interests needs to be prepared by inoculating already modified protein expression competent cell in 5ml to 25ml of autoclaved media. Media commonly used are LB, TB, etc. Starter culture needs to be incubated in appropriate temperature, such as 37 Celsius, along with well shaking overnight. On the same day, litters of media can be prepared. In the case of LB media, 25 grams of LB is required per liter of deionized water. The culture flasks are taped with aluminum foil and autoclaved. Before inoculation, the LB needs to be remained covered with aluminum foil to stay sterile. On the day of inoculation, the culture media needs to be cool to at least room temperature. Appropriate antibiotics needs to be added into the media with well shaking. Inoculation is done via adding 5-10ml of starter culture into each liter of culture. The media is then placed in a 37 degree shaker. It is important to keep track of the optical density of the inoculate culture. Desire optical density is 0.6. In the case of E. coli, such optical density is obtained after about 3 hours of inoculation. E. coli duplicate its amount every 20 minutes; however, in the presence of antibiotics, such duplication period might take longer. But a 3-hour inoculation period is safe. After 3 hours, the optical density of media should be carefully monitored. If the OD is too low, induction might not be sufficient; while if the OD is too high, we might obtain undesired protein, so an OD of 0.6 is desired. After the OD reaches 0.6, the media needs to be chilled on ice before induction. IPTG is commonly used to induce BL21 competent cell. 1mM of IPTG is sufficient for induction. The induction temperature might be different from inoculation temperature. Induction takes place over night.
On the second day, media is spun down into pellets. The pellets needs to be lysed via either a French or a microfluidizer depending on the amount of pellet available. French Press is usually good for a 2L culture lysis, while microfluidizer is a better choice for anything go beyond. The actual choice also depends on what is available and how soluble the pellets are in lysis buffer.
After the pellets are suspended in lysis buffer, lysozyme, DNase, and RNase are added, and inculated for at least 10 minutes. If inadequate amount of any of these are added, the pellets will appear sticky during lysis and lysis might be incomplete. After lysis, the sample is spun down to obtain the soluble fraction that contain our desired protein. It is crucial to keep the whole lysing process cold as some protein might precipitate in room temperature, or machine got heated up will cause a loss of protein. With the soluble fraction that contain our protein of interest, further purification can be done. Such as salting out, ion exchange, and affinity chromatography. Further purification might involve FPLC.
The appropriate lysis buffer is crucial in protein expression. Different plasmids express differently in different media. Temperature, and pH are also important factors to take into account.
References
edit- ↑ Gene expression
- ↑ Transcription (genetics)
- ↑ Beadle GW, Tatum EL (15 November 1941). "Genetic Control of Biochemical Reactions in Neurospora". PNAS 27 (11): 499–506.
- ↑ One gene-one enzyme hypothesis
- ↑ Operon
- ↑ http://en.wikipedia.org/w/index.php?title=Riboswitch&oldid=420979855
- ↑ Lac operon
- ↑ http://en.wikipedia.org/w/index.php?title=Lac_repressor&oldid=426589639
- ↑ http://en.wikipedia.org/w/index.php?title=Trp_operon&oldid=428240917
- ↑ Gene expression
- ↑ Plasmid
- ↑ Polymerase chain reaction
- ↑ Polymerase chain reaction
- ↑ Pingoud A, Jeltsch A (September 2001). "Structure and function of type II restriction endonucleases". Nucleic Acids Res. 29 (18): 3705–27. doi:10.1093/nar/29.18.3705. PMC 55916. PMID 11557805.
- ↑ Molecular Biology: Understanding the Genetic Revolution, by David P. Clark. Elsevier Academic Press, 2005. ISBN 0-12-175551-7.
- ↑ Goodsell DS (2002). "The molecular perspective: restriction endonucleases". Stem Cells. 20 (2): 190–1. doi:10.1634/stemcells.20-2-190. PMID 11897876.
- ↑ Roberts RJ (January 1980). "Restriction and modification enzymes and their recognition sequences". Nucleic Acids Res. 8 (1): r63–r80. doi:10.1093/nar/8.1.197-d. PMC 327257. PMID 6243774.
- ↑ R.J Roberts, 1988, Nucl Acids Res. 16(suppl):271 From p.213 Molecular Cell Biology 4th Edition by Lodish, Berk, Zipursky, Matsudaira, Baltimore and Darnell.
- ↑ "Stu I from Streptomyces tubercidicus". Sigma-Aldrich. Retrieved 2008-06-07.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - ↑ Shimotsu H, Takahashi H, Saito H (November 1980). "A new site-specific endonuclease StuI from Streptomyces tubercidicus". Gene. 11 (3–4): 219–25. doi:10.1016/0378-1119(80)90062-1. PMID 6260571.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Copeland NG, Jenkins NA, Court DL (2001). "Recombineering: a powerful new tool for mouse functional genomics". Nat. Rev. Genet. 2 (10): 769–79. doi:10.1038/35093556. PMID 11584293.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ↑ Lu JP, Beatty LK, Pinthus JH. (2008). "Dual expression recombinase based (DERB) single vector system for high throughput screening and verification of protein interactions in living cells". Nature Precedings.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ http://en.wikipedia.org/w/index.php?title=Reporter_gene&oldid=422027341