An Introduction to Molecular Biology/Cell Cycle
The cell cycle, or cell-division cycle (cdc), is the series of events that takes place in a cell leading to its division and duplication. In cells without a nucleus (prokaryotic), the cell cycle occurs via a process termed binary fission. In cells with a nucleus (eukaryotes), the cell cycle can be divided in two brief periods: interphase—during which the cell grows, accumulating nutrients needed for mitosis and duplicating its DNA—and the mitosis (M) phase, during which the cell splits itself into two distinct cells, often called "daughter cells". The cell-division cycle is a vital process by which a single-celled fertilized egg develops into a mature organism, as well as the process by which hair, skin, blood cells, and some internal organs are renewed.[1]
| |
A Wikibookian has nominated this page for cleanup. You can help make it better. Please review any relevant discussion. |
| A Wikibookian has nominated this page for cleanup. You can help make it better. Please review any relevant discussion. |
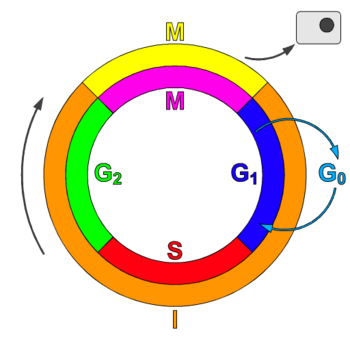
Phases of cell division
editThe cell cycle consists of four distinct phases: G1 (Gap1) phase, S phase (synthesis), G2 (Gap2) phase (collectively known as interphase) and M phase (mitosis). M (mitosis) phase is itself composed of two tightly coupled processes: mitosis, in which the cell's chromosomes are divided between the two daughter cells, and cytokinesis, in which the cell's cytoplasm divides in half forming distinct cells. Activation of each phase is dependent on the proper progression and completion of the previous one. Cells that have temporarily or reversibly stopped dividing are said to have entered a state of quiescence called G0 phase.[1]
G0 phase
editCells that have temporarily stopped dividing are said to have entered a state of quienscence, called G0 PHASE. G0 phase is viewed as either an extended G1 phase, where the cell is neither dividing nor preparing to divide, or a distinct quiescent stage that occurs outside of the cell cycle. G0 is sometimes referred to as a "post-mitotic" state, since cells in G0 are in a non-dividing phase outside of the cell cycle. Some types of cells, such as nerve and heart muscle cells, become post-mitotic when they reach maturity (i.e., when they are terminally differentiated) but continue to perform their main functions for the rest of the organism's life. Multinucleated muscle cells that do not undergo cytokinesis are also often considered to be in the G0 stage. On occasion, a distinction in terms is made between a G0 cell and a 'post-mitotic' cell (e.g., heart muscle cells and neurons), which will never enter the G1 phase, whereas other G0 cells may.[2]
G1 phase
editThe first phase of interphase is G1 phase, from the end of the previous Mitosis phase until the beginning of DNA replication is called G1 (G indicating gap). It is also called the growth phase. During this phase the biosynthetic activities of the cell, which had been considerably slowed down during M phase, resume at a high rate. This phase is marked by synthesis of various enzymes that are required in S phase, mainly those needed for DNA replication. Duration of G1 is highly variable, even among different cells of the same species.[1]
S phase
editInitiation of DNA replication is indication of S phase; when it is complete, all of the chromosomes have been replicated, at this time each chromosome has two (sister) chromatids. Thus, during this phase, the amount of DNA in the cell has effectively doubled, though the ploidy of the cell remains the same. Rates of RNA transcription and protein synthesis are very low during this phase. An exception to this is production of histone protein, which mostly occurs during the S phase.[1]
G2 phase
editAfter S phase or replication cell then enters the G2 phase, which lasts until the cell enters mitosis. Again, significant biosynthesis occurs during this phase, mainly involving the production of microtubules, which are required during the process of mitosis. Inhibition of protein synthesis during G2 phase prevents the cell from undergoing mitosis.
Analysis of Cell cycle
Cell cycle analysis is a method in cell biology that employs flow cytometry to distinguish cells in different phases of the cell cycle. Before analysis, the cells are permeabilised and treated with a fluorescent dye that stains DNA quantitatively, usually propidium iodide (PI). The fluorescence intensity of the stained cells at certain wavelengths will therefore correlate with the amount of DNA they contain. As the DNA content of cells duplicates during the S phase of the cell cycle, the relative amount of cells in the G0 phase and G1 phase (before S phase), in the S phase, and in the G2 phase and M phase (after S phase) can be determined, as the fluorescence of cells in the G2/M phase will be twice as high as that of cells in the G0/G1 phase. Cell cycle anomalies can be symptoms for various kinds of cell damage, for example DNA damage, which cause the cell to interrupt the cell cycle at certain checkpoints to prevent transformation into a cancer cell (carcinogenesis). Other possible reasons for anomalies include lack of nutrients, for example after serum deprivation. Cell cycle analysis was first described in 1969 at Los Alamos Scientific Laboratory by a group from the University of California[1], using the Feulgen staining technique. The first protocol for cell cycle analysis using propidium iodide staining was presented in 1975 by Awtar Krishan from Harvard Medical School and is still widely cited today.[3]
Mitosis
editMitosis is the process by which a eukaryotic cell separates the chromosomes in its nucleus into two identical sets in two nuclei. It is generally followed immediately by cytokinesis, which divides the nuclei, cytoplasm, organelles and cell membrane into two cells containing roughly equal shares of these cellular components. Mitosis and cytokinesis together define the mitotic (M) phase of the cell cycle - the division of the mother cell into two daughter cells, genetically identical to each other and to their parent cell. This accounts for approximately 10% of the cell cycle. Mitosis occurs exclusively in eukaryotic cells, but occurs in different ways in different species. For example, animals undergo an "open" mitosis, where the nuclear envelope breaks down before the chromosomes separate, while fungi such as Aspergillus nidulans and Saccharomyces cerevisiae (yeast) undergo a "closed" mitosis, where chromosomes divide within an intact cell nucleus. Prokaryotic cells, which lack a nucleus, divide by a process called binary fission. The process of mitosis is complex and highly regulated. The sequence of events is divided into phases, corresponding to the completion of one set of activities and the start of the next. These stages are prophase, prometaphase, metaphase, anaphase and telophase. During the process of mitosis the pairs of chromosomes condense and attach to fibers that pull the sister chromatids to opposite sides of the cell. The cell then divides in cytokinesis, to produce two identical daughter cells. Because cytokinesis usually occurs in conjunction with mitosis, "mitosis" is often used interchangeably with "M phase". However, there are many cells where mitosis and cytokinesis occur separately, forming single cells with multiple nuclei. This occurs most notably among the fungi and slime moulds, but is found in various different groups. Even in animals, cytokinesis and mitosis may occur independently, for instance during certain stages of fruit fly embryonic development. Errors in mitosis can either kill a cell through apoptosis or cause mutations that may lead to cancer.[4]
Prophase
Prophase, from the ancient Greek pro (before) and phase (stage), is a stage of mitosis in which the chromatin condenses (it becomes shorter and fatter) into a highly ordered structure called a chromosome in which the chromatin becomes visible. This process, called chromatin condensation, is mediated by the condensin complex. Since the genetic material has been duplicated in an earlier phase of the cell cycle, there are two identical copies of each chromosome in the cell. Identical chromosomes, called sister chromatids, are attached to each other at a DNA element present on every chromosome called the centromere. During prophase, giemsa staining can be applied to elicit G-banding in chromosomes. Prophase accounts for approximately 3% of the cell cycle's duration. An important organelle in mitosis is the centrosome, the microtubule organizing center in metazoans. During prophase, the two centrosomes, which replicate independently of mitosis, have their microtubule-activity increased due to the recruitment of γ-tubulin. The centrosomes will be pushed apart to opposite ends of the cell nucleus by the action of molecular motors acting on the microtubules. The nuclear envelope breaks down to allow the microtubules to reach the kinetochores on the chromosomes, marking the end of prophase. Prometaphase, the next step of mitosis, will see the chromosome being captured by the microtubules.[5]
Prophase in plant cells
In this first phase of mitosis, plant cells undergo a series of changes that is called puberty. In highly vacuolated plant cells, the contractile vacuole has to migrate into the center of the cell before mitosis can begin. This is achieved during the G2 phase of the cell cycle. A transverse sheet of cytoplasm bisects the cell along the future plane of cell division. Prophase in plant cells is preceded by a stage only found in plants, the formation of a ring of microtubules and actin filaments underneath the plasma membrane around the equatorial plane of the future mitotic spindle and predicting the position of cell plate fusion during telophase. During telophase in animal cells, a cleavage furrow forms. The preprophase band disappears during nuclear envelope disassembly and spindle formation in prometaphase despite contrary belief. The cells of higher plants lack centrioles. Instead, the nuclear envelope serves as a microtubule organising center. Spindle microtubules aggregate on the surface of the nuclear envelope during preprophase and prophase, forming the prophase spindle.[5]
Metaphase
Metaphase, from the ancient Greek meta (between) and phase (stage), is a stage of mitosis in the eukaryotic cell cycle in which condensed & highly coiled chromosomes, carrying genetic information, align in the middle of the cell before being separated into each of the two daughter cells. Metaphase accounts for approximately 4% of the cell cycle's duration. Preceded by events in prometaphase and followed by anaphase, microtubules formed in prophase have already found and attached themselves to kinetochores in metaphase. The centromeres of the chromosomes convene themselves on the metaphase plate (or equatorial plate), an imaginary line that is equidistant from the two centrosome poles. This even alignment is due to the counterbalance of the pulling powers generated by the opposing kinetochores, analogous to a tug of war between equally strong people. In certain types of cells, chromosomes do not line up at the metaphase plate and instead move back and forth between the poles randomly, only roughly lining up along the middleline. Early events of metaphase can coincide with the later events of prometaphase, as chromosomes with connected kinetochores will start the events of metaphase individually before other chromosomes with unconnected kinetochores that are still lingering in the events of prometaphase. One of the cell cycle checkpoints occurs during prometaphase and metaphase. Only after all chromosomes have become aligned at the metaphase plate, when every kinetochore is properly attached to a bundle of microtubules, does the cell enter anaphase. It is thought that unattached or improperly attached kinetochores generate a signal to prevent premature progression to anaphase, even if most of the kinetochores have been attached and most of the chromosomes have been aligned. Such a signal creates the mitotic spindle checkpoint. This would be accomplished by regulation of the anaphase-promoting complex, securin, and separase.[6]
Anaphase (ana (up) and phase (stage))
Anaphase begins abruptly with the regulated triggering of the metaphase-to-anaphase transition and accounts for approximately 1% of the cell cycle's duration. At this point, anaphase begins. This terminate activity by cleaving and inactivating the M-phase cyclin required for the function of M-phase cyclin dependent kinases (M-Cdks). It also cleaves securin, a protein that inhibits the protease known as separase. Separase then cleaves cohesin, a protein responsible for holding sister chromatids together. During early anaphase (or Anaphase A), the chromatids abruptly separate and move toward the spindle poles. This is achieved by the shortening of spindle microtubules, with forces mainly being exerted at the kinetochores. anaphase is when the chromatids separate from each other and move to opposite ends of the cell When the chromatids are fully separated, late anaphase (or Anaphase B) begins. This involves the polar microtubules elongating and sliding relative to each other to drive the spindle poles to opposite ends of the cell. Anaphase B drives the separation of sister centrosomes to opposite poles through three forces. Kinesin proteins that are attached to polar microtubules push the microtubules past one another. A second force involves the pulling of the microtubules by cortex-associated cytosolic dynein. The third force for chromosome separation involves the lengthening of the polar microtubules at their plus ends. These two processes were originally distinguished by their different sensitivities to drugs, and they are mechanically distinct. Early anaphase (Anaphase A) involves the shortening of kinetochore microtubules by depolymerization at their plus ends. During this process, a sliding collar allows chromatid movement. No motor protein is involved, as ATP depletion does not inhibit early anaphase. Late anaphase (Anaphase B) involves both the elongation of overlapping microtubules and the use of two distinct sets of motor proteins: one pulls overlapping microtubules past each other, and the other pulls astral microtubules that have attached to the cell cortex. The contributions of early anaphase and late anaphase to anaphase as a whole vary by cell type. In mammalian cells, late anaphase follows shortly after early anaphase and extends the spindle to approximately twice its metaphase length; by contrast, yeast and certain protozoans use late metaphase as the main means of chromosome separation and, in the process, can extend their spindles to up to 15 times the metaphase length.[7]
Cyclins
editCyclins are a Group of proteins that control the progression of cells through the cell cycle by activating Cyclin-dependent kinase (Cdk) enzymes.Cyclins were discovered by R. Timothy Hunt in 1982 while studying the cell cycle of sea urchins.
Types of Cyclins
editThere are several different cyclins that are active in different parts of the cell cycle and that cause the Cdk to phosphorylate different substrates.
There are two groups of cyclins:
G1/S cyclins – These cyclins are essential for the control of the cell cycle at the G1/S transition, Cyclin A / CDK2 – active in S phase. Cyclin D / CDK4, Cyclin D / CDK6, and Cyclin E / CDK2 – regulates transition from G1 to S phase.
G2/M cyclins – essential for the control of the cell cycle at the G2/M transition (mitosis). G2/M cyclins accumulate steadily during G2 and are abruptly destroyed as cells exit from mitosis (at the end of the M-phase). Cyclin B / CDK1 – regulates progression from G2 to M phase.
There are also several "orphan" cyclins for which no Cdk partner has been identified. For example, cyclin F is an orphan cyclin that is essential for G2/M transition.[8]
Cyclin dependent kinases (CDKs)
editCDKs are a family of protein kinases. CDKs are present in all known eukaryotes, and their regulatory function in the cell cycle has been evolutionarily conserved. CDKS are also involved in regulation of transcription, mRNA processing, and the differentiation of nerve cells. One interesting fact is that, yeast cells can proliferate normally when their CDK gene has been replaced with the homologous human gene. CDKs are relatively small proteins, with molecular weights ranging from 34 to 40 kDa, and contain little more than the kinase domain. CDK binds to a regulatory protein called a cyclin. Without cyclin, CDK has little kinase activity, only the cyclin-CDK complex is an active kinase. CDKs phosphorylate their substrates on serines and threonines, so they are serine-threonine kinases. The consensus sequence for the phosphorylation site in the amino acid sequence of a CDK substrate is [S/T*]PX[K/R], where S/T* is the phosphorylated serine or threonine, P is proline, X is any amino acid, K is lysine, and R is arginine.[9]
Table : Cyclin-dependent kinases that control the cell cycle in model organisms.[10]
| Species | Name | Original name | Size (amino acids) | Function |
|---|---|---|---|---|
| Saccharomyces cerevisiae | Cdk1 | Cdc28 | 298 | All cell-cycle stages |
| Schizosaccharomyces pombe | Cdk1 | Cdc2 | 297 | All cell-cycle stages |
| Drosophila melanogaster | Cdk1 | Cdc2 | 297 | M |
| Cdk2 | Cdc2c | 314 | G1/S, S, possibly M | |
| Cdk4 | Cdk4/6 | 317 | G1, promotes growth | |
| Xenopus laevis | Cdk1 | Cdc2 | 301 | M |
| Cdk2 | 297 | S, possibly M | ||
| Homo sapiens | Cdk1 | Cdc2 | 297 | M |
| Cdk2 | 298 | G1, S, possibly M | ||
| Cdk4 | 301 | G1 | ||
| Cdk6 | 326 | G1 |
Functions of cyclin and CDKs
editTwo key classes of regulatory molecules, cyclins and cyclin-dependent kinases (CDKs), determine a cell's progress through the cell cycle. Leland H. Hartwell, R. Timothy Hunt, and Paul M. Nurse won the 2001 Nobel Prize in Physiology or Medicine for their discovery of these central molecules. Many of the genes encoding cyclins and CDKs are conserved among all eukaryotes, but in general more complex organisms have more elaborate cell cycle control systems that incorporate more individual components. Many of the relevant genes were first identified by studying yeast, especially Saccharomyces cerevisiae; genetic nomenclature in yeast dubs many these genes cdc (for "cell division cycle") followed by an identifying number, e.g., cdc25 or cdc20.
Cyclins form the regulatory subunits and CDKs the catalytic subunits of an activated heterodimer; cyclins have no catalytic activity and CDKs are inactive in the absence of a partner cyclin. When activated by a bound cyclin, CDKs perform a common biochemical reaction called phosphorylation that activates or inactivates target proteins to orchestrate coordinated entry into the next phase of the cell cycle. Different cyclin-CDK combinations determine the downstream proteins targeted. CDKs are constitutively expressed in cells whereas cyclins are synthesised at specific stages of the cell cycle, in response to various molecular signals.
Upon receiving a pro-mitotic extracellular signal, G1 cyclin-CDK complexes become active to prepare the cell for S phase, promoting the expression of transcription factors that in turn promote the expression of S cyclins and of enzymes required for DNA replication. The G1 cyclin-CDK complexes also promote the degradation of molecules that function as S phase inhibitors by targeting them for ubiquitination. Once a protein has been ubiquitinated, it is targeted for proteolytic degradation by the proteasome. Active S cyclin-CDK complexes phosphorylate proteins that make up the pre-replication complexes assembled during G1 phase on DNA replication origins. The phosphorylation serves two purposes: to activate each already-assembled pre-replication complex, and to prevent new complexes from forming. This ensures that every portion of the cell's genome will be replicated once and only once. The reason for prevention of gaps in replication is fairly clear, because daughter cells that are missing all or part of crucial genes will die. However, for reasons related to gene copy number effects, possession of extra copies of certain genes is also deleterious to the daughter cells. Mitotic cyclin-CDK complexes, which are synthesized but inactivated during S and G2 phases, promote the initiation of mitosis by stimulating downstream proteins involved in chromosome condensation and mitotic spindle assembly. A critical complex activated during this process is a ubiquitin ligase known as the anaphase-promoting complex (APC), which promotes degradation of structural proteins associated with the chromosomal kinetochore. APC also targets the mitotic cyclins for degradation, ensuring that telophase and cytokinesis can proceed. Interphase: Interphase generally lasts at least 12 to 24 hours in mammalian tissue. During this period, the cell is constantly synthesizing RNA, producing protein and growing in size. By studying molecular events in cells, scientists have determined that interphase can be divided into 4 steps: Gap 0 (G0), Gap 1 (G1), S (synthesis) phase, Gap 2 (G2).
Cyclin D is the first cyclin produced in the cell cycle, in response to extracellular signals (e.g. growth factors). Cyclin D binds to existing CDK4, forming the active cyclin D-CDK4 complex. Cyclin D-CDK4 complex in turn phosphorylates the retinoblastoma susceptibility protein (Rb). The hyperphosphorylated Rb dissociates from the E2F/DP1/Rb complex (which was bound to the E2F responsive genes, effectively "blocking" them from transcription), activating E2F. Activation of E2F results in transcription of various genes like cyclin E, cyclin A, DNA polymerase, thymidine kinase, etc. Cyclin E thus produced binds to CDK2, forming the cyclin E-CDK2 complex, which pushes the cell from G1 to S phase (G1/S transition). Cyclin B along with cdc2 (cdc2 - fission yeasts (CDK1 - mammalia)) forms the cyclin B-cdc2 complex, which initiates the G2/M transition. Cyclin B-cdc2 complex activation causes breakdown of nuclear envelope and initiation of prophase, and subsequently, its deactivation causes the cell to exit mitosis.[1]
Disregulation of cell cycle
editA disregulation of the cell cycle components may lead to tumor formation. As mentioned above, some genes like the cell cycle inhibitors, RB, p53 etc., when they mutate, may cause the cell to multiply uncontrollably, forming a tumor. Although the duration of cell cycle in tumor cells is equal to or longer than that of normal cell cycle, the proportion of cells that are in active cell division (versus quiescent cells in G0 phase) in tumors is much higher than that in normal tissue. Thus there is a net increase in cell number as the number of cells that die by apoptosis or senescence remains the same. The cells which are actively undergoing cell cycle are targeted in cancer therapy as the DNA is relatively exposed during cell division and hence susceptible to damage by drugs or radiation. This fact is made use of in cancer treatment; by a process known as debulking, a significant mass of the tumor is removed which pushes a significant number of the remaining tumor cells from G0 to G1 phase (due to increased availability of nutrients, oxygen, growth factors etc.). Radiation or chemotherapy following the debulking procedure kills these cells which have newly entered the cell cycle. The fastest cycling mammalian cells in culture, crypt cells in the intestinal epithelium, have a cycle time as short as 9 to 10 hours. Stem cells in resting mouse skin may have a cycle time of more than 200 hours. Most of this difference is due to the varying length of G1, the most variable phase of the cycle. M and S do not vary much. In general, cells are most radiosensitive in late M and G2 phases and most resistant in late S. For cells with a longer cell cycle time and a significantly long G1 phase, there is a second peak of resistance late in G1 The pattern of resistance and sensitivity correlates with the level of sulfhydryl compounds in the cell. Sulfhydryls are natural radioprotectors and tend to be at their highest levels in S and at their lowest near mitosis.[1]
Cell cycle checkpoints
editThe G1/S checkpoint
editThe G1/S transition, more commonly known as the Start checkpoint in budding yeast (the restriction point in other organisms) regulates cell cycle commitment At this checkpoint, cells either arrest before DNA replication (due to limiting nutrients or a pheromone signal), prolong G1 (size control), or begin replication and progress through the rest of the cell cycle. The G1/S regulatory network or regulon in budding yeast includes the G1 cyclins Cln1, Cln2 and Cln3, Cdc28 (Cdk1), the transcription factors SBF and MBF, and the transcriptional inhibitor Whi5.[11] Cln3 interacts with Cdk1 to initiate the sequence of events by phosphorylating a large number of targets, including SBF, MBF and Whi5. Phosphorylation of Whi5 causes it to translocate out of the nucleus, preventing it from inhibiting SBF and MBF. Active SBF/MBF drive the G1/S transition by turning on the B-type cyclins and initiating DNA replication, bud formation and spindle body duplication. Moreover, SBF/MBF drives expression of Cln1 and Cln2, which can also interact with Cdk1 to promote phosphorylation of its targets.
This G1/S switch was initially thought to function as a linear sequence of events starting with Cln3 and ending in S phase.[12] However, the observation that any one of the Clns was sufficient to activate the regulon indicated that Cln1 and Cln2 might be able to engage positive feedback to activate their own transcription. This would result in a continuously accelerating cycle that could act as an irreversible bistable trigger. Skotheim et al. used single-cell measurements in budding yeast to show that this positive feedback does indeed occur. A small amount of Cln3 induces Cln1/2 expression and then the feedback loop takes over, leading to rapid and abrupt exit of Whi5 from the nucleus and consequently coherent expression of G1/S regulon genes. In the absence of coherent gene expression, cells take longer to exit G1 and a significant fraction even arrest before S phase, highlighting the importance of positive feedback in sharpening the G1/S switch.
The G1/S cell cycle checkpoint controls the passage of eukaryotic cells from the first gap phase, G1, into the DNA synthesis phase, S. In this switch in mammalian cells, there are two cell cycle kinases that help to control the checkpoint: cell cycle kinases CDK4/6-cyclin D and CDK2-cyclin E. The transcription complex that includes Rb and E2F is important in controlling this checkpoint. In the first gap phase, the Rb-HDAC repressor complex binds to the E2F-DP1 transcription factors, therefore inhibiting the downstream transcription. The phosphorylation of Rb by CDK4/6 and CDK2 dissociates the Rb-repressor complex and serves as an on/off switch for the cell cycle. Once Rb is phosphorylated, the inhibition is released on the E2F transcriptional activity. This allows for the transcription of S phase genes encoding for proteins that amplify the G1 to S phase switch.[13]
Many different stimuli apply checkpoint controls including TGFb, DNA damage, contact inhibition, replicative senescence, and growth factor withdrawal. The first four act by inducing members of the INK4 or Kip/Cip families of cell cycle kinase inhibitors. TGFb inhibits the transcription of Cdc25A, a phosphatase that activates the cell cycle kinases, and growth factor withdrawal activates GSK3b, which phosphorylates cyclin D. This leads to its rapid ubiquitination.[14]
The G2/M checkpoint
editThis transition is commenced by E2F-mediated transcription of cyclin A, forming the cyclin A-Cdk2 complex. This is useful in regulating events in prophase. In order to proceed past prophase, the cyclin B-Cdk1 complex (first discovered as MPF or M-phase promoting factor) is activated by Cdc 25, a protein phosphatase1. As mitosis starts, the nuclear envelope disintegrates, chromosomes condense and become visibile, and the cells prepares for division. The Cyclin B-Cdk1 activation results in nuclear envelope breakdown, which is a characteristic of the initiation of mitosis.It is evident that the cyclin A and B complexes with Cdks help regulate mitotic events at the G2/M transition.[15]
As mentioned above, entry into mitosis is controlled by the Cyclin B-Cdk1 complex (first discovered as MPF or M-phase promoting factor; Cdk1 is also known as Cdc2 in fission yeast and Cdc28 in budding yeast). This complex forms an element of an interesting regulatory circuit in which Cdk1 can phosphorylate and activate its activator, the phosphatase Cdc25 (positive feedback), and phosphorylate and inactivate its inactivator, the kinase Wee1 (double-negative feedback). It was suggested that this circuit could act as a bistable trigger[16] with one stable steady state in G2 (Cdk and Cdc25 off, Wee1 on) and a second stable steady state in M phase (Cdk and Cdc25 active, Wee1 off). Once cells are in mitosis, Cyclin B-Cdk1 activates the Anaphase-promoting complex (APC), which in turn inactivates Cyclin B-Cdk1 by degrading Cyclin B, eventually leading to exit from mitosis. Coupling the bistable Cdk1 response function to the negative feedback from the APC could generate what is known as a relaxation oscillator,[17] with sharp spikes of Cdk1 activity triggering robust mitotic cycles. However, in a relaxation oscillator, the control parameter moves slowly relative to the system’s response dynamics which may be an accurate representation of mitotic entry, but not necessarily mitotic exit.
It is necessary to inactivate the cyclin B-Cdk1 complex in order to exit the mitotic stage of the cell cycle. The cells can then return to the first gap phase G1 and wait until the cycle proceeds yet again.
In 2003 Pomerening et al. provided strong evidence for this hypothesis by demonstrating hysteresis and bistability in the activation of Cdk1 in the cytoplasmic extracts of Xenopus oocytes.[17] They first demonstrated a discontinuous sharp response of Cdk1 to changing concentrations of non-destructible Cyclin B (to decouple the Cdk1 response network from APC-mediated negative feedback). However, such a response would be consistent with both a monostable, ultransensitive transition and a bistable transition. To distinguish between these two possibilities, they measured the steady-state levels of active Cdk1 in response to changing cyclin levels, but in two separate experiments, one starting with an interphase extract and one starting with an extract already in mitosis. At intermediate concentrations of cyclin they found two steady-state concentrations of active Cdk1. Which of the two steady states was occupied depended on the history of the system, i.e.whether they started with interphase or mitotic extract, effectively demonstrating hysteresis and bistability.
In the same year, Sha et al.[18] independently reached the same conclusion revealing the hysteretic loop also using Xenopus laevis egg extracts. In this article, three predictions of the Novak-Tyson model were tested in an effort to conclude that hysteresis is the driving force for “cell-cycle transitions into and out of mitosis”. The predictions of the Novak-Tyson model are generic to all saddle-node bifurcations. Saddle-node bifurcations are extremely useful bifurcations in an imperfect world because they help describe biological systems which are not perfect. The first prediction was that the threshold concentration of cyclin to enter mitosis is higher than the threshold concentration of cyclin to exit mitosis, and this was confirmed by supplementing cycling egg extracts with non-degradable cyclin B and measuring the activation and inactivation threshold after the addition of cycloheximide (CHX), which is a protein synthesis inhibitor. Furthermore, the second prediction of the Novak-Tyson model was also validated: unreplicated deoxyribonucleic acid, or DNA, increases the threshold concentration of cyclin that is required to enter mitosis. In order to arrive at this conclusion, cytostatic factor released extracts were supplemented with CHX, APH (a DNA polymerase inhibitor), or both, and non-degradable cyclin B was added. The third and last prediction that was tested and proven true in this article was that the rate of Cdc2 activation slows down near the activation threshold concentration of cyclin. These predictions and experiments demonstrate the toggle-like switching behavior that can be described by hysteresis in a dynamical system.[19]
Metaphase-anaphase checkpoint
editIn the transition from Spindle checkpoint|metaphase to anaphase, it is crucial that sister chromatids are properly and simultaneously separated to opposite ends of the cell. Separation of sister-chromatids is initially strongly inhibited to prevent premature separation in late mitosis, but this inhibition is relieved through destruction of the inhibitory elements by the anaphase-promoting complex (APC) once sister-chromatid bi-orientation is achieved. One of these inhibitory elements is securin, which prevents the destruction of cohesin, the complex that holds the sister-chromatids together, by binding the protease separase which targets Scc1, a subunit of the cohesin complex, for destruction. In this system, the phosphatase Cdc14 can remove an inhibitory phosphate from securin, thereby facilitating the destruction of securin by the APC, releasing separase. As shown by Uhlmann et al., during the attachment of chromosomes to the mitotic spindle the chromatids remain paired because cohesion between the sisters prevents separation.[20] Cohesion is established during DNA replication and depends on cohesin, which is a multisubunit complex composed of Scc1, Scc3, Smc2, and Smc3. In yeast at the metaphase-to-anaphase transition, Scc1 dissociates from the chromosomes and the sister chromatids separate. This action is controlled by the Esp1 protein, which is tightly bound by the anaphase inhibitor Pds1 that is destroyed by the anaphase-promoting complex. In order to verify that Esp1 does play a role in regulating Scc1 chromosome association, cell strains were arrested in G1 with an alpha factor. These cells stayed in arrest during the development. Esp1-1 mutant cells were used and the experiment was repeated, and Scc1 successfully bound to the chromosomes and remained associated even after the synthesis was terminated. This was crucial in showing that with Esp1, Scc1 is hindered in its ability to become stably associated with chromosomes during G1, and Esp1 can in fact directly remove Scc1 from chromosomes.[13]
It has been shown by Holt et al.[21] that separase activates Cdc14, which in turn acts on securin, thus creating a positive feedback loop that increases the sharpness of the metaphase to anaphase transition and coordination of sister-chromatid separation.[21] Holt et al. probed the basis for the effect of positive feedback in securin phosphophorlyation by using mutant 'securin' strains of yeast, and testing how changes in the phosphoregulation of securin affects the synchrony of sister chromatid separation. Their results indicate that interfering with this positive securin-separase-cdc14 loop decreases sister chromatid separation synchrony. This positive feedback can hypothetically generate bistability in the transition to anaphase, causing the cell to make the irreversible decision to separate sister-chromatids.
Cell division in fission yeast
editThe fission yeast is a single-celled fungus with simple, fully characterized genome and a rapid growth rate. It has long since been used in brewing, baking and molecular genetics. S. pombe is a rod-shaped cell, approximately 3 µm in diameter, that grows entirely by elongation at the ends. After mitosis, division occurs by the formation of a septum, or cell plate, that cleaves the cell at its midpoint.
The central events of cell reproduction are chromosome duplication, which takes place in S (Synthetic) phase, followed by chromosome segregation and nuclear division (mitosis) and cell division (cytokinesis), which are collectively called M (Mitotic) phase.G1 is the gap between M and S phases, and G2 is the gap between S and M phases. In the budding yeast, the G2 phase is particularly extended, and cytokinesis (daughter-cell segregation) does not happen until a new S (Synthetic) phase is launched.
Fission yeast governs mitosis by mechanisms that are similar to those in multicellular animals. It normally proliferates in a haploid state. When starved, cells of opposite mating types (P and M) fuse to form a diploid zygote that immediately enters meiosis to generate four haploid spores. When conditions improve, these spores germinate to produce proliferating haploid cells.[22]
Facts to be remembered
edit| State | Phase | Abbreviation | Description |
|---|---|---|---|
| quiescent/ senescent |
Gap 0 | G0 | A resting phase where the cell has left the cycle and has stopped dividing. |
| Interphase | Gap 1 | G1 | Cells increase in size in Gap 1. The G1 checkpoint control mechanism ensures that everything is ready for DNA synthesis. |
| Synthesis | S | DNA replication occurs during this phase. | |
| Gap 2 | G2 | During the gap between DNA synthesis and mitosis, the cell will continue to grow. The G2 checkpoint control mechanism ensures that everything is ready to enter the M (mitosis) phase and divide. | |
| Cell division | Mitosis | M | Cell growth stops at this stage and cellular energy is focused on the orderly division into two daughter cells. A checkpoint in the middle of mitosis (Metaphase Checkpoint) ensures that the cell is ready to complete cell division. |
Stages of mitosis [4]
Real mitotic cells can be visualized through the microscope by staining them with fluorescent antibodies and dyes. These light micrographs are included below.
-
Early prophase: Nonkinetochore microtubules, shown as green strands, have established a matrix around the degrading nucleus, in blue. The green nodules are the centrosomes.
-
Early prometaphase: The nuclear membrane has just degraded, allowing the microtubules to quickly interact with the kinetochores on the chromosomes, which have just condensed.
-
Late metaphase: The centrosomes have moved to the poles of the cell and have established the mitotic spindle. The chromosomes, in light blue, have all assembled at the metaphase plate, except for one.
-
Anaphase: Lengthening nonkinetochore microtubules push the two sets of chromosomes further apart.
References
edit- ↑ a b c d e f Cell cycle
- ↑ G0 phase
- ↑ Cell cycle analysis
- ↑ a b Mitosis
- ↑ a b Prophase
- ↑ Metaphase
- ↑ Anaphase
- ↑ Cyclins
- ↑ Cyclin-dependent kinase
- ↑ Morgan, David O. (2007). The Cell Cycle: Principles of Control. London: New Science Press, 1st ed.
- ↑ Skotheim, J.M.; Di Talia, S.; Siggia, E.D.; Cross, F.R. (2008), "Positive feedback of G1 cyclins ensures coherent cell cycle entry", Nature, 454 (7202): 291, retrieved 2009-12-11
- ↑ Stuart, D.; Wittenberg, C. (1995), "CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells." (PDF), Genes & development, 9 (22): 2780, retrieved 2009-12-11
- ↑ a b Biochemical switches in the cell cycle
- ↑ Harper JW. A phosphorylation-driven ubiquitination switch for cell cycle control. TrendsCell Biol. 2002 Mar;12(3):104-7. PMID 11859016
- ↑ Biochemical switches in the cell cycle
- ↑ Novak, B.; Tyson, J.J. (1993), "Numerical analysis of a comprehensive model of M-phase control in Xenopus oocyte extracts and intact embryos", Journal of Cell Science, 106 (4): 1153, retrieved 2009-12-11
- ↑ a b Pomerening, J. R., E. D. Sontag, et al. (2003). "Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2." Nat Cell Biol 5(4): 346-351.
- ↑ Sha, W.; Moore, J.; Chen, K.; Lassaletta, A.D.; Yi, C.S.; Tyson, J.J.; Sible, J.C. (2003), "Hysteresis drives cell-cycle transitions in Xenopus laevis egg extracts", Proceedings of the National Academy of Sciences, 100 (3): 975, retrieved 2009-12-11
- ↑ Cooper, G. (2000), “The Cell: A Molecular Approach.”, retrieved 2010-11-21
- ↑ Uhlmann F.; Lottspeich F.; Nasmyth K. (1999), “Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesion subunit Scc1,” Nature 400: 37-42, retrieved 2010-9-25
- ↑ a b Holt, L. J., A. N. Krutchinsky, et al. (2008). "Positive feedback sharpens the anaphase switch." Nature 454(7202): 353-357.
- ↑ Schizosaccharomyces pombe